1AW9
 
 | | STRUCTURE OF GLUTATHIONE S-TRANSFERASE III IN APO FORM | | Descriptor: | CADMIUM ION, GLUTATHIONE S-TRANSFERASE III | | Authors: | Neuefeind, T, Huber, R, Reinemer, P, Knaeblein, J. | | Deposit date: | 1997-10-13 | | Release date: | 1998-10-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cloning, sequencing, crystallization and X-ray structure of glutathione S-transferase-III from Zea mays var. mutin: a leading enzyme in detoxification of maize herbicides.
J.Mol.Biol., 274, 1997
|
|
1BLI
 
 | | BACILLUS LICHENIFORMIS ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Machius, M, Declerck, N, Huber, R, Wiegand, G. | | Deposit date: | 1998-01-07 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activation of Bacillus licheniformis alpha-amylase through a disorder-->order transition of the substrate-binding site mediated by a calcium-sodium-calcium metal triad.
Structure, 6, 1998
|
|
1CTS
 
 | |
1CHM
 
 | | ENZYMATIC MECHANISM OF CREATINE AMIDINOHYDROLASE AS DEDUCED FROM CRYSTAL STRUCTURES | | Descriptor: | CARBAMOYL SARCOSINE, CREATINE AMIDINOHYDROLASE | | Authors: | Hoeffken, H.W, Knof, S.H, Bartlett, P.A, Huber, R, Moellering, H, Schumacher, G. | | Deposit date: | 1993-07-19 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Enzymatic mechanism of creatine amidinohydrolase as deduced from crystal structures.
J.Mol.Biol., 214, 1990
|
|
1CL1
 
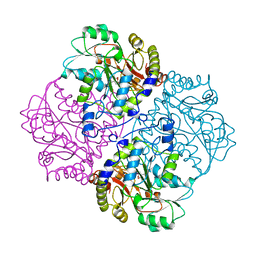 | |
1CL2
 
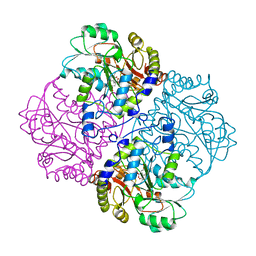 | |
1DHP
 
 | | DIHYDRODIPICOLINATE SYNTHASE | | Descriptor: | DIHYDRODIPICOLINATE SYNTHASE, POTASSIUM ION | | Authors: | Mirwaldt, C, Korndoerfer, I, Huber, R. | | Deposit date: | 1995-02-09 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of dihydrodipicolinate synthase from Escherichia coli at 2.5 A resolution.
J.Mol.Biol., 246, 1995
|
|
2JDX
 
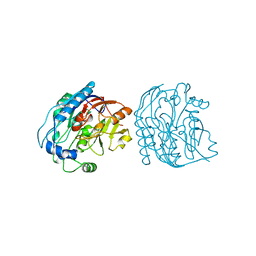 | |
1VLB
 
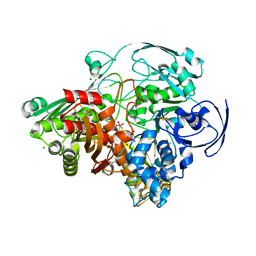 | | STRUCTURE REFINEMENT OF THE ALDEHYDE OXIDOREDUCTASE FROM DESULFOVIBRIO GIGAS AT 1.28 A | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ALDEHYDE OXIDOREDUCTASE, CHLORIDE ION, ... | | Authors: | Rebelo, J.M, Dias, J.M, Huber, R, Moura, J.J.G, Romao, M.J. | | Deposit date: | 2004-07-20 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure refinement of the aldehyde oxidoreductase from Desulfovibrio gigas (MOP) at 1.28 A
J.Biol.Inorg.Chem., 6, 2001
|
|
1TGB
 
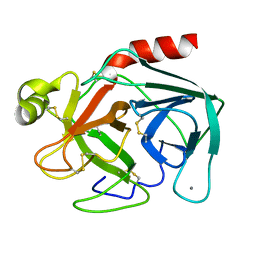 | | CRYSTAL STRUCTURE OF BOVINE TRYPSINOGEN AT 1.8 ANGSTROMS RESOLUTION. II. CRYSTALLOGRAPHIC REFINEMENT, REFINED CRYSTAL STRUCTURE AND COMPARISON WITH BOVINE TRYPSIN | | Descriptor: | CALCIUM ION, TRYPSINOGEN | | Authors: | Bode, W, Fehlhammer, H, Huber, R. | | Deposit date: | 1979-03-07 | | Release date: | 1979-06-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of bovine trypsinogen at 1-8 A resolution. II. Crystallographic refinement, refined crystal structure and comparison with bovine trypsin.
J.Mol.Biol., 111, 1977
|
|
1TYW
 
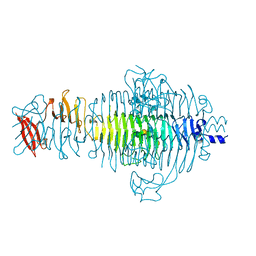 | | STRUCTURE OF TAILSPIKE-PROTEIN | | Descriptor: | TAILSPIKE PROTEIN, alpha-D-galactopyranose-(1-2)-[alpha-D-Tyvelopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-alpha-L-rhamnopyranose-(1-3)-[alpha-D-glucopyranose-(1-4)]alpha-D-galactopyranose-(1-2)-[alpha-D-Tyvelopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-alpha-L-rhamnopyranose | | Authors: | Steinbacher, S, Huber, R. | | Deposit date: | 1996-07-26 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1TYX
 
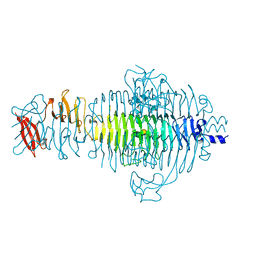 | | TITLE OF TAILSPIKE-PROTEIN | | Descriptor: | TAILSPIKE PROTEIN, alpha-D-galactopyranose-(1-2)-[alpha-D-Abequopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-alpha-L-rhamnopyranose-(1-3)-alpha-D-galactopyranose-(1-2)-[alpha-D-Abequopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-alpha-L-rhamnopyranose | | Authors: | Steinbacher, S, Huber, R. | | Deposit date: | 1996-07-26 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
3GPJ
 
 | | Crystal structure of the yeast 20S proteasome in complex with syringolin B | | Descriptor: | N-{[(1S)-2-methyl-1-{[(5S,8S)-5-(1-methylethyl)-2,7-dioxo-1,6-diazacyclododec-3-en-8-yl]carbamoyl}propyl]carbamoyl}-L-valine, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Huber, R, Kaiser, M. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic and structural studies on syringolin A and B reveal critical determinants of selectivity and potency of proteasome inhibition
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1TYU
 
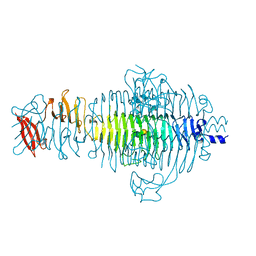 | | STRUCTURE OF TAILSPIKE-PROTEIN | | Descriptor: | TAILSPIKE PROTEIN, alpha-D-galactopyranose-(1-2)-[alpha-D-Tyvelopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-alpha-L-rhamnopyranose-(1-3)-alpha-D-galactopyranose-(1-2)-[alpha-D-Tyvelopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-alpha-L-rhamnopyranose | | Authors: | Steinbacher, S, Huber, R. | | Deposit date: | 1996-07-26 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1TYV
 
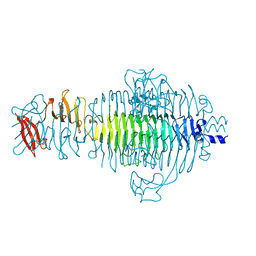 | | STRUCTURE OF TAILSPIKE-PROTEIN | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Steinbacher, S, Huber, R. | | Deposit date: | 1996-07-26 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1C7N
 
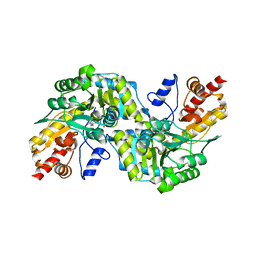 | | CRYSTAL STRUCTURE OF CYSTALYSIN FROM TREPONEMA DENTICOLA CONTAINS A PYRIDOXAL 5'-PHOSPHATE COFACTOR | | Descriptor: | CYSTALYSIN, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Krupka, H.I, Huber, R, Holt, S.C, Clausen, T. | | Deposit date: | 2000-03-16 | | Release date: | 2000-07-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of cystalysin from Treponema denticola: a pyridoxal 5'-phosphate-dependent protein acting as a haemolytic enzyme.
EMBO J., 19, 2000
|
|
1C7O
 
 | | CRYSTAL STRUCTURE OF CYSTALYSIN FROM TREPONEMA DENTICOLA CONTAINS A PYRIDOXAL 5'-PHOSPHATE-L-AMINOETHOXYVINYLGLYCINE COMPLEX | | Descriptor: | (2E,3E)-4-(2-aminoethoxy)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, CYSTALYSIN | | Authors: | Krupka, H.I, Huber, R, Holt, S.C, Clausen, T. | | Deposit date: | 2000-03-16 | | Release date: | 2000-07-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of cystalysin from Treponema denticola: a pyridoxal 5'-phosphate-dependent protein acting as a haemolytic enzyme.
EMBO J., 19, 2000
|
|
1D02
 
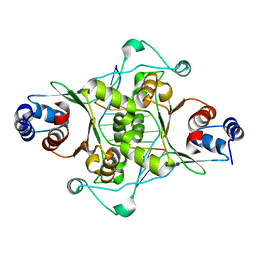 | | CRYSTAL STRUCTURE OF MUNI RESTRICTION ENDONUCLEASE IN COMPLEX WITH COGNATE DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*TP*TP*GP*GP*C)-3'), TYPE II RESTRICTION ENZYME MUNI | | Authors: | Deibert, M, Grazulis, S, Janulaitis, A, Siksnys, V, Huber, R. | | Deposit date: | 1999-09-08 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MunI restriction endonuclease in complex with cognate DNA at 1.7 A resolution.
EMBO J., 18, 1999
|
|
1DGJ
 
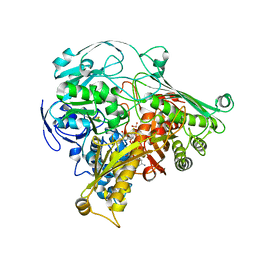 | | CRYSTAL STRUCTURE OF THE ALDEHYDE OXIDOREDUCTASE FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 | | Descriptor: | ALDEHYDE OXIDOREDUCTASE, FE2/S2 (INORGANIC) CLUSTER, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Rebelo, J.M, Macieira, S, Dias, J.M, Huber, R, Romao, M.J. | | Deposit date: | 1999-11-24 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Gene sequence and crystal structure of the aldehyde oxidoreductase from Desulfovibrio desulfuricans ATCC 27774.
J.Mol.Biol., 297, 2000
|
|
1DD4
 
 | | Crystal structure of ribosomal protein l12 from thermotoga maritim | | Descriptor: | 50S RIBOSOMAL PROTEIN L7/L12, HEXATANTALUM DODECABROMIDE | | Authors: | Wahl, M.C, Bourenkov, G.P, Bartunik, H.D, Huber, R. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Flexibility, conformational diversity and two dimerization modes in complexes of ribosomal protein L12.
Embo J., 19, 2000
|
|
1DD3
 
 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L12 FROM THERMOTOGA MARITIMA | | Descriptor: | 50S RIBOSOMAL PROTEIN L7/L12 | | Authors: | Wahl, M.C, Bourenkov, G.P, Bartunik, H.D, Huber, R. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexibility, conformational diversity and two dimerization modes in complexes of ribosomal protein L12.
EMBO J., 19, 2000
|
|
1DMG
 
 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L4 | | Descriptor: | CITRIC ACID, RIBOSOMAL PROTEIN L4 | | Authors: | Worbs, M, Huber, R, Wahl, M.C. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of ribosomal protein L4 shows RNA-binding sites for ribosome incorporation and feedback control of the S10 operon.
EMBO J., 19, 2000
|
|
2ABZ
 
 | | Crystal structure of C19A/C43A mutant of leech carboxypeptidase inhibitor in complex with bovine carboxypeptidase A | | Descriptor: | Carboxypeptidase A1, Metallocarboxypeptidase inhibitor, ZINC ION | | Authors: | Arolas, J.L, Popowicz, G.M, Bronsoms, S, Aviles, F.X, Huber, R, Holak, T.A, Ventura, S. | | Deposit date: | 2005-07-18 | | Release date: | 2006-01-31 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Study of a major intermediate in the oxidative folding of leech carboxypeptidase inhibitor: contribution of the fourth disulfide bond
J.Mol.Biol., 352, 2005
|
|
2NSM
 
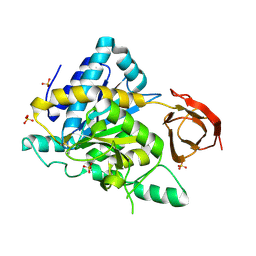 | | Crystal structure of the human carboxypeptidase N (Kininase I) catalytic domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carboxypeptidase N catalytic chain, SULFATE ION | | Authors: | Keil, C, Maskos, K, Than, M, Hoopes, J.T, Huber, R, Tan, F, Deddish, P.A, Erdoes, E.G, Skidgel, R.A, Bode, W. | | Deposit date: | 2006-11-05 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the human carboxypeptidase N (kininase I) catalytic domain
J.Mol.Biol., 366, 2007
|
|
1ELQ
 
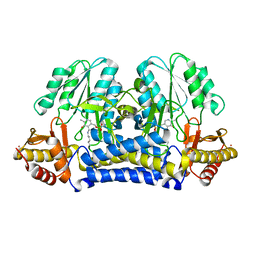 | | CRYSTAL STRUCTURE OF THE CYSTINE C-S LYASE C-DES | | Descriptor: | L-CYSTEINE/L-CYSTINE C-S LYASE, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Clausen, T, Kaiser, J.T, Steegborn, C, Huber, R, Kessler, D. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cystine C-S lyase from Synechocystis: stabilization of cysteine persulfide for FeS cluster biosynthesis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
