6Q8V
 
 | |
1ME3
 
 | |
1ME4
 
 | |
3CWB
 
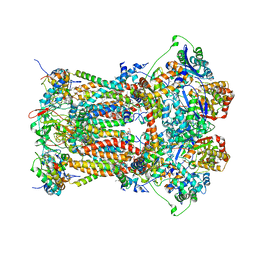 | | Chicken Cytochrome BC1 Complex inhibited by an iodinated analogue of the polyketide Crocacin-D | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, AZIDE ION, CARDIOLIPIN, ... | | Authors: | Huang, L, Cromartie, T, Viner, R, Crowley, P.J, Berry, E.A. | | Deposit date: | 2008-04-21 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | The role of molecular modeling in the design of analogues of the fungicidal natural products crocacins A and D.
Bioorg.Med.Chem., 16, 2008
|
|
4CS1
 
 | |
6L62
 
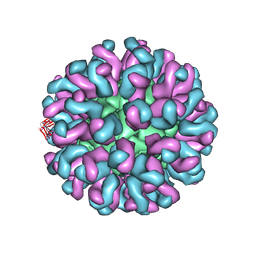 | | Neutralization mechanism of a monoclonal antibody targeting a porcine circovirus type 2 Cap protein conformational epitope | | Descriptor: | Capsid protein, Heavy chain of Fab fragment, Light chain of Fab fragment | | Authors: | Sun, Z, Huang, L, Xia, D, Wei, Y, Sun, E, Zhu, H, Bian, H, Wu, H, Feng, L, Wang, J, Liu, C. | | Deposit date: | 2019-10-25 | | Release date: | 2020-02-12 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Neutralization Mechanism of a Monoclonal Antibody Targeting a Porcine Circovirus Type 2 Cap Protein Conformational Epitope.
J.Virol., 94, 2020
|
|
6LM3
 
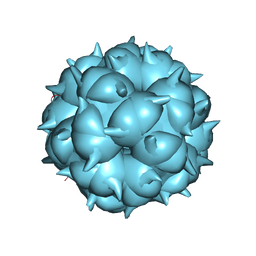 | | Neutralization mechanism of a monoclonal antibody targeting a porcine circovirus type 2 Cap protein conformational epitope | | Descriptor: | Capsid protein | | Authors: | Sun, Z, Huang, L, Xia, D, Wei, Y, Sun, E, Zhu, H, Bian, H, Wu, H, Feng, L, Wang, J, Liu, C. | | Deposit date: | 2019-12-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Neutralization Mechanism of a Monoclonal Antibody Targeting a Porcine Circovirus Type 2 Cap Protein Conformational Epitope.
J.Virol., 94, 2020
|
|
1BCC
 
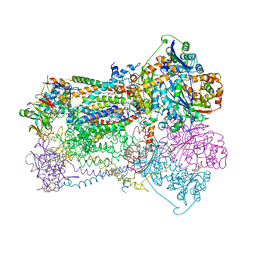 | | CYTOCHROME BC1 COMPLEX FROM CHICKEN | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.-I, Kim, K.K, Hung, L.-W, Crofts, A.R, Berry, E.A, Kim, S.-H. | | Deposit date: | 1998-03-23 | | Release date: | 1998-08-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Electron transfer by domain movement in cytochrome bc1.
Nature, 392, 1998
|
|
1DJ0
 
 | | THE CRYSTAL STRUCTURE OF E. COLI PSEUDOURIDINE SYNTHASE I AT 1.5 ANGSTROM RESOLUTION | | Descriptor: | CHLORIDE ION, PSEUDOURIDINE SYNTHASE I | | Authors: | Foster, P.G, Huang, L, Santi, D.V, Stroud, R.M. | | Deposit date: | 1999-11-30 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I.
Nat.Struct.Biol., 7, 2000
|
|
5WVZ
 
 | | The crystal structure of Cren7 mutant L28F in complex with dsDNA | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2016-12-30 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
5WVY
 
 | | The crystal structure of Cren7 mutant L28V in complex with dsDNA | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2016-12-29 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
5WVW
 
 | | The crystal structure of Cren7 mutant L28A in complex with dsDNA | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2016-12-29 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
5WWC
 
 | | The crystal structure of Cren7 mutant L28M in complex with dsDNA | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2016-12-31 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
2BCC
 
 | | STIGMATELLIN-BOUND CYTOCHROME BC1 COMPLEX FROM CHICKEN | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.I, Kim, K.K, Hung, L.W, Crofts, A.R, Berry, E.A, Kim, S.H. | | Deposit date: | 1998-09-18 | | Release date: | 1999-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
2OJU
 
 | | X-ray structure of complex of human cyclophilin J with cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE-LIKE 3 | | Authors: | Xia, Z, Huang, L. | | Deposit date: | 2007-01-14 | | Release date: | 2008-01-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting Cyclophilin J, a Novel Peptidyl-Prolyl Isomerase, Can Induce Cellular G1/S Arrest and Repress the Growth of Hepatocellular Carcinoma
To be Published
|
|
6NBU
 
 | |
6U7V
 
 | | xRRM structure of spPof8 | | Descriptor: | NITRATE ION, Protein pof8 | | Authors: | Kim, J.-K, Hu, X, Yu, C, Jun, H.-I, Liu, J, Sankaran, B, Huang, L, Qiao, F. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Quality-Control Mechanism for Telomerase RNA Folding in the Cell.
Cell Rep, 33, 2020
|
|
3H1I
 
 | | Stigmatellin and antimycin bound cytochrome bc1 complex from chicken | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-METHYL-BUTYRIC ACID 3-(3-FORMYLAMINO-2-HYDROXY-BENZOYLAMINO)-8-HEPTYL-2,6-DIMETHYL-4,9-DIOXO-[1,5]DIOXONAN-7-YL ESTER, CARDIOLIPIN, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.I, Kim, K.K, Hung, L.W, Crofts, A.R, Berry, E.A, Kim, S.H. | | Deposit date: | 2009-04-12 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
3H1H
 
 | | Cytochrome bc1 complex from chicken | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, CYTOCHROME C1, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.I, Kim, K.K, Hung, L.W, Crofts, A.R, Berry, E.A, Kim, S.H. | | Deposit date: | 2009-04-12 | | Release date: | 2009-04-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
4R56
 
 | | Crystal structure of Sulfolobus Cren7-dsDNA(GTGATCAC) complex | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3') | | Authors: | Zhang, Z.F, Gong, Y, Chen, Y.Y, Li, H.B, Huang, L. | | Deposit date: | 2014-08-20 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into the interaction between Cren7 and DNA: the role of loop beta 3-beta 4
Extremophiles, 19, 2015
|
|
4R55
 
 | | The crystal structure of a Cren7 mutant protein GR and dsDNA complex | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3') | | Authors: | Zhang, Z.F, Gong, Y, Chen, Y.Y, Li, H.B, Huang, L. | | Deposit date: | 2014-08-20 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the interaction between Cren7 and DNA: the role of loop beta 3-beta 4
Extremophiles, 19, 2015
|
|
3LWI
 
 | | Crystal structure of Cren7-dsDNA complex | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3') | | Authors: | Zhang, Z.F, Gong, Y, Guo, L, Jiang, T, Huang, L. | | Deposit date: | 2010-02-23 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the interaction of the crenarchaeal chromatin protein Cren7 with DNA
Mol.Microbiol., 76, 2010
|
|
3LWH
 
 | | Crystal structure of Cren7-dsDNA complex | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3') | | Authors: | Zhang, Z.F, Gong, Y, Guo, L, Jiang, T, Huang, L. | | Deposit date: | 2010-02-23 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the interaction of the crenarchaeal chromatin protein Cren7 with DNA
Mol.Microbiol., 76, 2010
|
|
8FNI
 
 | | Cryo-EM structure of RNase-treated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNF
 
 | | Cryo-EM structure of RNase-untreated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
