2G50
 
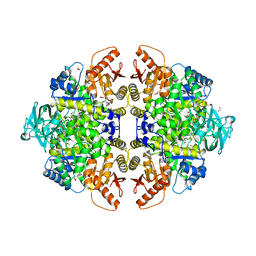 | | The location of the allosteric amino acid binding site of muscle pyruvate kinase. | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ALANINE, ... | | Authors: | Holyoak, T, Williams, R, Fenton, A.W. | | Deposit date: | 2006-02-22 | | Release date: | 2006-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Differentiating a Ligand's Chemical Requirements for Allosteric Interactions from Those for Protein Binding. Phenylalanine Inhibition of Pyruvate Kinase.
Biochemistry, 45, 2006
|
|
4IMA
 
 | | The structure of C436M-hLPYK in complex with Citrate/Mn/ATP/Fru-1,6-BP | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE, ... | | Authors: | Zhang, B, Holyoak, T, Fenton, A.W, Tang, Q.L, Prasannan, C.B, Deng, J.P. | | Deposit date: | 2013-01-02 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Energetic Coupling between an Oxidizable Cysteine and the Phosphorylatable N-Terminus of Human Liver Pyruvate Kinase.
Biochemistry, 52, 2013
|
|
6P5O
 
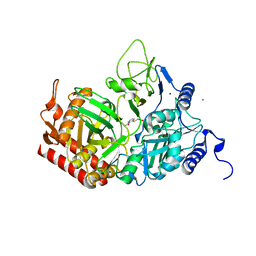 | |
8DDL
 
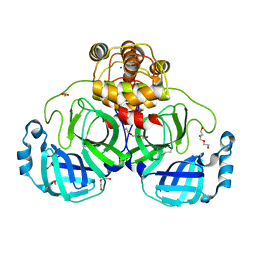 | | SARS-CoV-2 Main Protease (Mpro) H163A Mutant Apo Structure | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ORF1a polyprotein, ... | | Authors: | Tran, N, McLeod, M.J, Kalyaanamoorthy, S, Ganesan, A, Holyoak, T. | | Deposit date: | 2022-06-18 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun, 14, 2023
|
|
8DD6
 
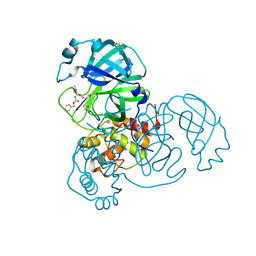 | | SARS-CoV-2 Main Protease (Mpro) H163A Mutant in Complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, DIMETHYL SULFOXIDE, ORF1a polyprotein | | Authors: | Tran, N, McLeod, M.J, Kalyaanamoorthy, S, Ganesan, A, Holyoak, T. | | Deposit date: | 2022-06-17 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun, 14, 2023
|
|
3H09
 
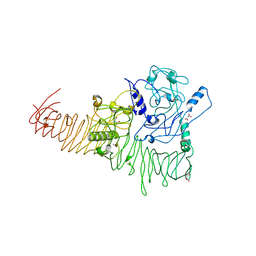 | | The structure of Haemophilus influenzae IgA1 protease | | Descriptor: | ACETATE ION, Immunoglobulin A1 protease, MALONIC ACID, ... | | Authors: | Johnson, T.A, Qiu, J, Plaut, A.G, Holyoak, T. | | Deposit date: | 2009-04-08 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active-Site Gating Regulates Substrate Selectivity in a Chymotrypsin-Like Serine Protease The Structure of Haemophilus influenzae Immunoglobulin A1 Protease.
J.Mol.Biol., 389, 2009
|
|
8SG6
 
 | | SARS-CoV-2 Main Protease (Mpro) H163A Mutant Reduced with 20mM TCEP | | Descriptor: | 3C-like proteinase nsp5, GLYCEROL | | Authors: | Tran, N, McLeod, M.J, Barwell, S, Kalyaanamoorthy, S, Ganesan, A, Holyoak, T. | | Deposit date: | 2023-04-11 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun, 14, 2023
|
|
3R9I
 
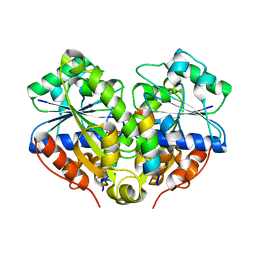 | | 2.6A resolution structure of MinD complexed with MinE (12-31) peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division topological specificity factor, Septum site-determining protein minD | | Authors: | Lovell, S, Battaile, K.P, Park, K.-T, Wu, W, Holyoak, T, Lutkenhaus, J. | | Deposit date: | 2011-03-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Min Oscillator Uses MinD-Dependent Conformational Changes in MinE to Spatially Regulate Cytokinesis.
Cell(Cambridge,Mass.), 146, 2011
|
|
1XQL
 
 | | Effect of a Y265F Mutant on the Transamination Based Cycloserine Inactivation of Alanine Racemase | | Descriptor: | (5-HYDROXY-4-{[(3-HYDROXYISOXAZOL-4-YL)AMINO]METHYL}-6-METHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, (R)-4-AMINO-ISOXAZOLIDIN-3-ONE, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Fenn, T.D, Holyoak, T, Stamper, G.F, Ringe, D. | | Deposit date: | 2004-10-12 | | Release date: | 2005-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effect of a Y265F Mutant on the Transamination-Based Cycloserine Inactivation of Alanine Racemase
Biochemistry, 44, 2005
|
|
1XQK
 
 | | Effect of a Y265F Mutant on the Transamination Based Cycloserine Inactivation of Alanine Racemase | | Descriptor: | (5-HYDROXY-4-{[(3-HYDROXYISOXAZOL-4-YL)AMINO]METHYL}-6-METHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Alanine racemase | | Authors: | Fenn, T.D, Holyoak, T, Stamper, G.F, Ringe, D. | | Deposit date: | 2004-10-12 | | Release date: | 2005-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Effect of a Y265F Mutant on the Transamination-Based Cycloserine Inactivation of Alanine Racemase
Biochemistry, 44, 2005
|
|
4YWD
 
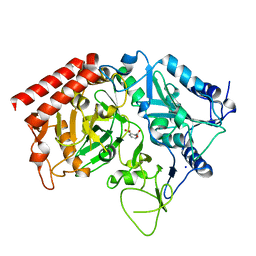 | | Structure of rat cytosolic pepck in complex with 2,3-Pyridine dicarboxylic acid | | Descriptor: | MANGANESE (II) ION, Phosphoenolpyruvate carboxykinase, cytosolic [GTP], ... | | Authors: | Balan, M.D, Johnson, T.A, Mcleod, M.J, Lotosky, W.R, Holyoak, T. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition and Allosteric Regulation of Monomeric Phosphoenolpyruvate Carboxykinase by 3-Mercaptopicolinic Acid.
Biochemistry, 54, 2015
|
|
4YW8
 
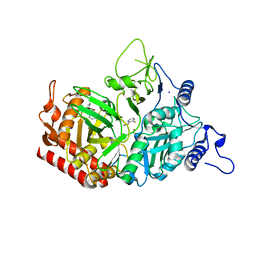 | | Structure of rat cytosolic pepck in complex with 3-mercaptopicolinic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-sulfanylpyridine-2-carboxylic acid, MANGANESE (II) ION, ... | | Authors: | Balan, M.D, Johnson, T.A, Mcleod, M.J, Lotosky, W.R, Holyoak, T. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Inhibition and Allosteric Regulation of Monomeric Phosphoenolpyruvate Carboxykinase by 3-Mercaptopicolinic Acid.
Biochemistry, 54, 2015
|
|
4YW9
 
 | | Structure of rat cytosolic pepck in complex with 3-mercaptopicolinic acid and GTP | | Descriptor: | 3-sulfanylpyridine-2-carboxylic acid, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Balan, M.D, Johnson, T.A, Mcleod, M.J, Lotosky, W.R, Holyoak, T. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition and Allosteric Regulation of Monomeric Phosphoenolpyruvate Carboxykinase by 3-Mercaptopicolinic Acid.
Biochemistry, 54, 2015
|
|
4YWB
 
 | | Structure of rat cytosolic pepck in complex with 3-mercaptopicolinic acid and oxalic acid | | Descriptor: | 3-sulfanylpyridine-2-carboxylic acid, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Balan, M.D, Johnson, T.A, Lotosky, W.R, Mcleod, M.J, Holyoak, T. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibition and Allosteric Regulation of Monomeric Phosphoenolpyruvate Carboxykinase by 3-Mercaptopicolinic Acid.
Biochemistry, 54, 2015
|
|
8E1U
 
 | |
2ID4
 
 | |
7L3M
 
 | | PEPCK MMQX structure 40ms post-mixing with oxaloacetic acid | | Descriptor: | CARBON DIOXIDE, GUANOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Clinger, J.A, Moreau, D.W, McLeod, M.J, Holyoak, T, Thorne, R.E. | | Deposit date: | 2020-12-18 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Millisecond mix-and-quench crystallography (MMQX) enables time-resolved studies of PEPCK with remote data collection.
Iucrj, 8, 2021
|
|
7L3V
 
 | | PEPCK MMQX structure 120ms post-mixing with oxaloacetic acid | | Descriptor: | CARBON DIOXIDE, GUANOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Clinger, J.A, Moreau, D.W, McLeod, M.J, Holyoak, T, Thorne, R.E. | | Deposit date: | 2020-12-18 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Millisecond mix-and-quench crystallography (MMQX) enables time-resolved studies of PEPCK with remote data collection.
Iucrj, 8, 2021
|
|
7L36
 
 | | PEPCK steady-state structure with Mn and GTP | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Clinger, J.A, Moreau, D.W, McLeod, M.J, Holyoak, T, Thorne, R.E. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Millisecond mix-and-quench crystallography (MMQX) enables time-resolved studies of PEPCK with remote data collection.
Iucrj, 8, 2021
|
|
3DT4
 
 | |
3DTB
 
 | |
3DT7
 
 | |
3DT2
 
 | |
5FH2
 
 | | The structure of rat cytosolic PEPCK variant E89Q in complex with GTP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Johnson, T.A, Holyoak, T. | | Deposit date: | 2015-12-21 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Utilization of Substrate Intrinsic Binding Energy for Conformational Change and Catalytic Function in Phosphoenolpyruvate Carboxykinase.
Biochemistry, 55, 2016
|
|
5FH5
 
 | |
