5APY
 
 | |
5APU
 
 | | Sequence IANKEDKAD inserted between GCN4 adaptors - Structure A9b black | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, UREA | | Authors: | Hartmann, M.D, Mendler, C.T, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
5APP
 
 | | Actinobacillus actinomycetemcomitans OMP100 residues 133-198 fused to GCN4 adaptors | | Descriptor: | CHLORIDE ION, General control protein GCN4,GENERAL CONTROL PROTEIN GCN4, OUTER MEMBRANE PROTEIN 100,General control protein GCN4 | | Authors: | Hartmann, M.D, Ridderbusch, O, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
5AMK
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in multiple conformations, hexagonal crystal form | | Descriptor: | CEREBLON ISOFORM 4, DIMETHYL SULFOXIDE, S-Thalidomide, ... | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-03-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Dynamics of the Cereblon Ligand Binding Domain.
Plos One, 10, 2015
|
|
5APS
 
 | |
5AMH
 
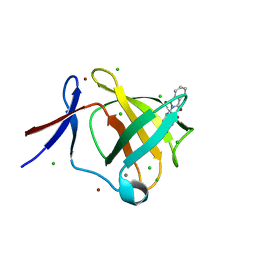 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thalidomide, trigonal crystal form | | Descriptor: | CALCIUM ION, CEREBLON ISOFORM 4, CHLORIDE ION, ... | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-03-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Dynamics of the Cereblon Ligand Binding Domain.
Plos One, 10, 2015
|
|
5AMI
 
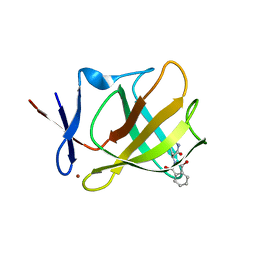 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thalidomide, Wash I structure | | Descriptor: | CEREBLON ISOFORM 4, S-Thalidomide, ZINC ION | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-03-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Dynamics of the Cereblon Ligand Binding Domain.
Plos One, 10, 2015
|
|
5APX
 
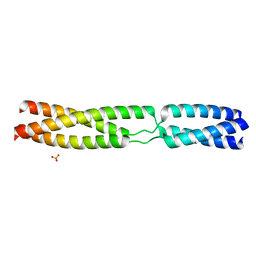 | | Sequence MATKDDIAN inserted between GCN4 adaptors - Structure T9(6) | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PHOSPHATE ION | | Authors: | Hartmann, M.D, Mendler, C.T, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
5APW
 
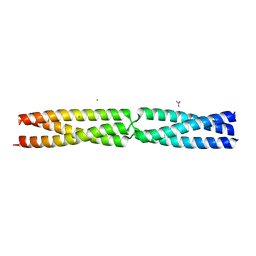 | | Sequence MATKDD inserted between GCN4 adaptors - Structure T6 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GENERAL CONTROL PROTEIN GCN4, ... | | Authors: | Hartmann, M.D, Mendler, C.T, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
5APT
 
 | |
5APV
 
 | |
5APQ
 
 | |
5AMJ
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thalidomide, Wash II structure | | Descriptor: | CEREBLON ISOFORM 4, PHOSPHATE ION, S-Thalidomide, ... | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-03-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Dynamics of the Cereblon Ligand Binding Domain.
Plos One, 10, 2015
|
|
5APZ
 
 | | Thermosinus carboxydivorans Nor1 Tcar0761 residues 68-101 and 191-211 fused to GCN4 adaptors | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, NOR1 TCAR0761 | | Authors: | Hartmann, M.D, Deiss, S, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
6Y06
 
 | |
6Y07
 
 | |
7NY0
 
 | |
4C47
 
 | |
3H7Z
 
 | |
3H7X
 
 | |
6R0V
 
 | |
6R0U
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with compound 5a and hydrolysis product | | Descriptor: | 3-azanyl-2-[[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]carbamoyl]benzoic acid, 4-azanyl-2-[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]isoindole-1,3-dione, CHLORIDE ION, ... | | Authors: | Heim, C, Hartmann, M.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De-Novo Design of Cereblon (CRBN) Effectors Guided by Natural Hydrolysis Products of Thalidomide Derivatives.
J.Med.Chem., 62, 2019
|
|
6R0S
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with compound 4a and hydrolysis product | | Descriptor: | 2-[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]-4-nitro-isoindole-1,3-dione, 2-[[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]carbamoyl]-6-nitro-benzoic acid, CEREBLON ISOFORM 4, ... | | Authors: | Heim, C, Hartmann, M.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | De-Novo Design of Cereblon (CRBN) Effectors Guided by Natural Hydrolysis Products of Thalidomide Derivatives.
J.Med.Chem., 62, 2019
|
|
6R0Q
 
 | |
6R11
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with compound 5b | | Descriptor: | 5-azanyl-2-[(3~{S})-2,5-bis(oxidanylidene)pyrrolidin-3-yl]isoindole-1,3-dione, CHLORIDE ION, Cereblon isoform 4, ... | | Authors: | Heim, C, Hartmann, M.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | De-Novo Design of Cereblon (CRBN) Effectors Guided by Natural Hydrolysis Products of Thalidomide Derivatives.
J.Med.Chem., 62, 2019
|
|
