8EM4
 
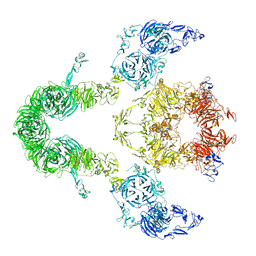 | | Cryo-EM structure of LRP2 at pH 7.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Beenken, A, Cerutti, G, Brasch, J, Fitzpatrick, A.W, Barasch, J, Shapiro, L. | | 登録日 | 2022-09-26 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-03-08 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | Structures of LRP2 reveal a molecular machine for endocytosis.
Cell, 186, 2023
|
|
3DJ3
 
 | |
3DIW
 
 | | c-terminal beta-catenin bound TIP-1 structure | | 分子名称: | Tax1-binding protein 3, decameric peptide form Catenin beta-1 | | 著者 | Shen, Y. | | 登録日 | 2008-06-21 | | 公開日 | 2008-10-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis of beta-Catenin Recognition by Tax-interacting Protein-1
J.Mol.Biol., 384, 2008
|
|
3DJ1
 
 | | crystal structure of TIP-1 wild type | | 分子名称: | SULFATE ION, Tax1-binding protein 3 | | 著者 | Shen, Y. | | 登録日 | 2008-06-21 | | 公開日 | 2008-10-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis of beta-Catenin Recognition by Tax-interacting Protein-1
J.Mol.Biol., 384, 2008
|
|
3Q1J
 
 | | Crystal structure of tudor domain 1 of human PHD finger protein 20 | | 分子名称: | PHD finger protein 20, UNKNOWN ATOM OR ION | | 著者 | Tempel, W, Li, Z, Wernimont, A.K, Chao, X, Bian, C, Lam, R, Crombet, L, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2010-12-17 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structures of the Tudor domains of human PHF20 reveal novel structural variations on the Royal Family of proteins.
Febs Lett., 586, 2012
|
|
3QII
 
 | | Crystal structure of tudor domain 2 of human PHD finger protein 20 | | 分子名称: | PHD finger protein 20, UNKNOWN ATOM OR ION | | 著者 | Li, Z, Tempel, W, Wernimont, A.K, Chao, X, Bian, C, Lam, R, Crombet, L, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2011-01-27 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of the Tudor domains of human PHF20 reveal novel structural variations on the Royal Family of proteins.
Febs Lett., 586, 2012
|
|
5TZY
 
 | | GPR40 in complex with AgoPAM AP8 and partial agonist MK-8666 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3R)-3-cyclopropyl-3-[(2R)-2-(1-{(1S)-1-[5-fluoro-2-(trifluoromethoxy)phenyl]ethyl}piperidin-4-yl)-3,4-dihydro-2H-1-benzopyran-7-yl]-2-methylpropanoic acid, (5aR,6S,6aS)-3-({2',6'-dimethyl-4'-[3-(methylsulfonyl)propoxy][1,1'-biphenyl]-3-yl}methoxy)-5,5a,6,6a-tetrahydrocyclopropa[4,5]cyclopenta[1,2-c]pyridine-6-carboxylic acid, ... | | 著者 | Lu, J, Byrne, N, Patel, S, Sharma, S, Soisson, S.M. | | 登録日 | 2016-11-22 | | 公開日 | 2017-06-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.22 Å) | | 主引用文献 | Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5TZR
 
 | | GPR40 in complex with partial agonist MK-8666 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (5aR,6S,6aS)-3-({2',6'-dimethyl-4'-[3-(methylsulfonyl)propoxy][1,1'-biphenyl]-3-yl}methoxy)-5,5a,6,6a-tetrahydrocyclopropa[4,5]cyclopenta[1,2-c]pyridine-6-carboxylic acid, Free fatty acid receptor 1,Endolysin,Free fatty acid receptor 1, ... | | 著者 | Lu, J, Byrne, N, Patel, S, Sharma, S, Soisson, S.M. | | 登録日 | 2016-11-22 | | 公開日 | 2017-06-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6VU2
 
 | | M1214_N1 Fab structure | | 分子名称: | M1214 N1 Fab heavy chain, M1214 N1 Fab light chain | | 著者 | Pan, R, Kong, X. | | 登録日 | 2020-02-14 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | VSV-Displayed HIV-1 Envelope Identifies Broadly Neutralizing Antibodies Class-Switched to IgG and IgA.
Cell Host Microbe, 27, 2020
|
|
6VY2
 
 | |
2OSL
 
 | | Crystal structure of Rituximab Fab in complex with an epitope peptide | | 分子名称: | B-lymphocyte antigen CD20, heavy chain of the Rituximab Fab fragment,heavy chain of the Rituximab Fab fragment, light chain of the Rituximab Fab fragment,light chain of the Rituximab Fab fragment | | 著者 | Du, J, Zhong, C, Ding, J. | | 登録日 | 2007-02-06 | | 公開日 | 2007-04-10 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for recognition of CD20 by therapeutic antibody Rituximab
J.Biol.Chem., 282, 2007
|
|
2PVX
 
 | | NMR and X-ray Analysis of Structural Additivity in Metal Binding Site-Swapped Hybrids of Rubredoxin | | 分子名称: | Rubredoxin, ZINC ION | | 著者 | Wang, L, LeMaster, D.M, Hernandez, G, Li, H. | | 登録日 | 2007-05-10 | | 公開日 | 2007-12-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.04 Å) | | 主引用文献 | NMR and X-ray analysis of structural additivity in metal binding site-swapped hybrids of rubredoxin
Bmc Struct.Biol., 7, 2007
|
|
3L1N
 
 | | Crystal structure of Mp1p ligand binding domain 2 complexd with palmitic acid | | 分子名称: | Cell wall antigen, PALMITIC ACID | | 著者 | Liao, S, Tung, E.T, Zheng, W, Chong, K, Xu, Y, Bartlam, M, Rao, Z, Yuen, K.Y. | | 登録日 | 2009-12-14 | | 公開日 | 2010-01-05 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystal structure of the Mp1p ligand binding domain 2 reveals its function as a fatty acid-binding protein.
J.Biol.Chem., 285, 2010
|
|
3F7O
 
 | | Crystal structure of Cuticle-Degrading Protease from Paecilomyces lilacinus (PL646) | | 分子名称: | (MSU)(ALA)(ALA)(PRO)(VAL), CALCIUM ION, Serine protease | | 著者 | Liang, L, Lou, Z, Meng, Z, Rao, Z, Zhang, K. | | 登録日 | 2008-11-10 | | 公開日 | 2009-11-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The crystal structures of two cuticle-degrading proteases from nematophagous fungi and their contribution to infection against nematodes.
Faseb J., 24, 2010
|
|
3F7M
 
 | | Crystal structure of apo Cuticle-Degrading Protease (ver112) from Verticillium psalliotae | | 分子名称: | Alkaline serine protease ver112 | | 著者 | Liang, L, Lou, Z, Ye, F, Meng, Z, Rao, Z, Zhang, K. | | 登録日 | 2008-11-09 | | 公開日 | 2009-11-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The crystal structures of two cuticle-degrading proteases from nematophagous fungi and their contribution to infection against nematodes.
Faseb J., 24, 2010
|
|
5CX3
 
 | | Crystal structure of FYCO1 LIR in complex with LC3A | | 分子名称: | FYVE and coiled-coil domain-containing protein 1, GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3A | | 著者 | Cheng, X, Pan, L. | | 登録日 | 2015-07-28 | | 公開日 | 2016-08-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis of FYCO1 and MAP1LC3A interaction reveals a novel binding mode for Atg8-family proteins.
Autophagy, 12, 2016
|
|
3G6L
 
 | |
3G6M
 
 | | crystal structure of a chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with a potent inhibitor caffeine | | 分子名称: | CAFFEINE, Chitinase | | 著者 | Gan, Z, Yang, J, Lou, Z, Rao, Z, Zhang, K.-Q. | | 登録日 | 2009-02-06 | | 公開日 | 2010-02-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure and mutagenesis analysis of chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with the inhibitor caffeine
Microbiology, 156, 2010
|
|
5EOA
 
 | | Crystal structure of OPTN E50K mutant and TBK1 complex | | 分子名称: | Optineurin, Serine/threonine-protein kinase TBK1 | | 著者 | Li, F, Xie, X, Liu, J, Pan, L. | | 登録日 | 2015-11-10 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.503 Å) | | 主引用文献 | Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins.
Nat Commun, 7, 2016
|
|
7CPV
 
 | | Cryo-EM structure of 80S ribosome from mouse testis | | 分子名称: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | 著者 | Huo, Y.G, He, X, Jiang, T, Qin, Y, Guo, X.J, Sha, J.H. | | 登録日 | 2020-08-08 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-01-11 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | A male germ-cell-specific ribosome controls male fertility.
Nature, 612, 2022
|
|
6J56
 
 | |
6M17
 
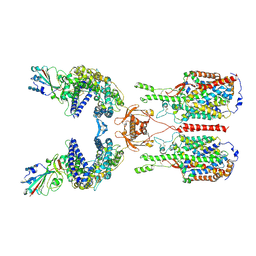 | | The 2019-nCoV RBD/ACE2-B0AT1 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yan, R.H, Zhang, Y.Y, Li, Y.N, Xia, L, Guo, Y.Y, Zhou, Q. | | 登録日 | 2020-02-24 | | 公開日 | 2020-03-11 | | 最終更新日 | 2021-03-10 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2.
Science, 367, 2020
|
|
4TKQ
 
 | | Native-SAD phasing for YetJ from Bacillus Subtilis | | 分子名称: | CALCIUM ION, CHLORIDE ION, Uncharacterized protein YetJ | | 著者 | Liu, Q, Chang, Y, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2014-05-27 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8025 Å) | | 主引用文献 | Multi-crystal native SAD analysis at 6 keV.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5EP6
 
 | | The crystal structure of NAP1 in complex with TBK1 | | 分子名称: | 5-azacytidine-induced protein 2, GLYCEROL, Serine/threonine-protein kinase TBK1 | | 著者 | Li, F, Xie, X, Liu, J, Pan, L. | | 登録日 | 2015-11-11 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.451 Å) | | 主引用文献 | Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins.
Nat Commun, 7, 2016
|
|
5EOF
 
 | | Crystal structure of OPTN NTD and TBK1 CTD complex | | 分子名称: | Optineurin, Serine/threonine-protein kinase TBK1 | | 著者 | Li, F, Xie, X, Liu, J, Pan, L. | | 登録日 | 2015-11-10 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins.
Nat Commun, 7, 2016
|
|
