2LSC
 
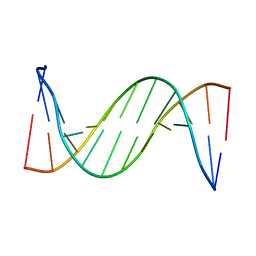 | | Solution structure of 2'F-ANA and ANA self-complementary duplex | | Descriptor: | DNA (5'-D(*(CFL)P*(GFL)P*(CFL)P*(GFL)P*(A5O)P*(A5O)P*(UAR)P*(UAR)P*(CFL)P*(GFL)P*(CFL)P*(GFL))-3') | | Authors: | Martin-Pintado, N, Yahyaee, M, Campos, R, Noronha, A, Wilds, C, Damha, M, Gonzalez, C. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The solution structure of double helical arabino nucleic acids (ANA and 2'F-ANA): effect of arabinoses in duplex-hairpin interconversion.
Nucleic Acids Res., 40, 2012
|
|
8BQY
 
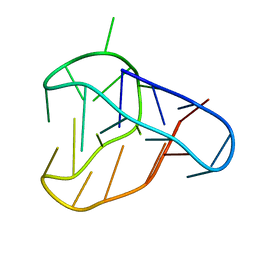 | | An i-motif domain able to undergo pH-dependent conformational transitions (acidic structure) | | Descriptor: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*(DNR)P*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*(DNR)P*CP*GP*T)-3') | | Authors: | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2022-11-22 | | Release date: | 2023-02-22 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
8BV6
 
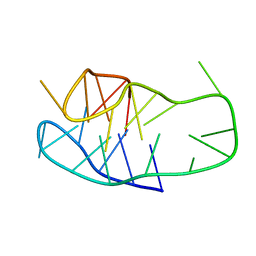 | | An i-motif domain able to undergo pH-dependent conformational transitions (neutral structure) | | Descriptor: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*CP*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*CP*CP*GP*T)-3') | | Authors: | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2022-12-01 | | Release date: | 2023-02-22 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
6FC9
 
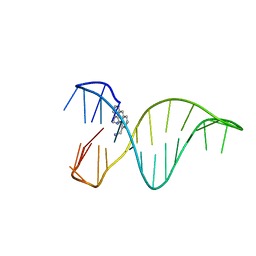 | | The 1,8-bis(aminomethyl)anthracene and Quadruplex-duplex junction complex | | Descriptor: | DNA (27-MER), [8-(azaniumylmethyl)anthracen-1-yl]methylazanium | | Authors: | Santana, A, Serrano, I, Montalvillo-Jimenez, L, Corzana, F, Bastida, A, Jimenez-Barbero, J, Gonzalez, C, Asensio, J.L. | | Deposit date: | 2017-12-20 | | Release date: | 2019-04-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | De Novo Design of Selective Quadruplex-Duplex Junction Ligands and Structural Characterisation of Their Binding Mode: Targeting the G4 Hot-Spot.
Chemistry, 2020
|
|
2HK4
 
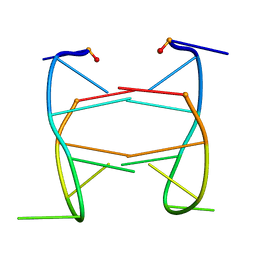 | |
2JO5
 
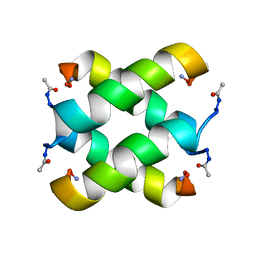 | | Tetrameric structure of KIA7F peptide | | Descriptor: | KIA7F | | Authors: | Lopez de la Osa, J, Gonzalez, C, Laurents, D.V, Chakrabartty, A, Bateman, D.A. | | Deposit date: | 2007-02-21 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Getting specificity from simplicity in putative proteins from the prebiotic earth.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2K40
 
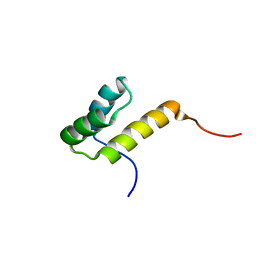 | |
2JO4
 
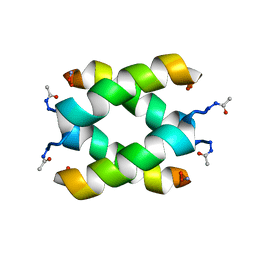 | | Tetrameric structure of KIA7 peptide | | Descriptor: | KIA7 | | Authors: | Lopez de la Osa, J, Gonzalez, C, Laurents, D.V, Chakrabartty, A, Bateman, D.A. | | Deposit date: | 2007-02-21 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Getting specificity from simplicity in putative proteins from the prebiotic earth.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2JML
 
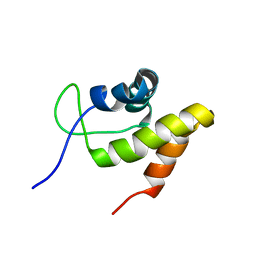 | | Solution structure of the N-terminal domain of CarA repressor | | Descriptor: | DNA BINDING DOMAIN/TRANSCRIPTIONAL REGULATOR | | Authors: | Jimenez, M, Padmanabhan, S, Gonzalez, C, Perez-Marin, M.C, Elias-Arnanz, M, Murillo, F.J, Rico, M. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural basis for operator and antirepressor recognition by Myxococcus xanthus CarA repressor.
Mol.Microbiol., 63, 2007
|
|
2K8Z
 
 | | Dimeric solution structure of the DNA loop d(TCGTTGCT) | | Descriptor: | 5'-D(*TP*CP*GP*TP*TP*GP*CP*T)-3' | | Authors: | Viladoms, J, Escaja, N, Frieden, M, Gomez-Pinto, I, Pedroso, E, Gonzalez, C. | | Deposit date: | 2008-09-29 | | Release date: | 2009-04-28 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Self-association of short DNA loops through minor groove C:G:G:C tetrads.
Nucleic Acids Res., 37, 2009
|
|
2K97
 
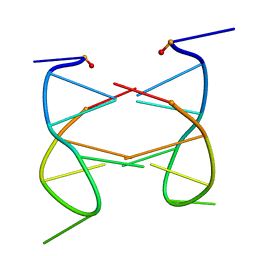 | | Dimeric solution structure of the cyclic octamer d(pCGCTCCGT) | | Descriptor: | 5'-D(P*CP*GP*CP*TP*CP*CP*GP*T)-3' | | Authors: | Viladoms, J, Escaja, N, Frieden, M, Gomez-Pinto, I, Pedroso, E, Gonzalez, C. | | Deposit date: | 2008-09-30 | | Release date: | 2009-04-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Self-association of short DNA loops through minor groove C:G:G:C tetrads.
Nucleic Acids Res., 37, 2009
|
|
2K90
 
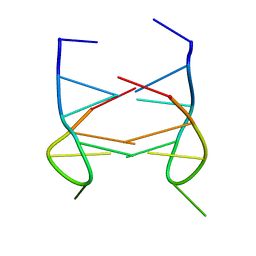 | | Dimeric solution structure of the DNA loop d(TGCTTCGT) | | Descriptor: | 5'-D(*TP*GP*CP*TP*TP*CP*GP*T)-3' | | Authors: | Viladoms, J, Escaja, N, Frieden, M, Gomez-Pinto, I, Pedroso, E, Gonzalez, C. | | Deposit date: | 2008-09-29 | | Release date: | 2009-04-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Self-association of short DNA loops through minor groove C:G:G:C tetrads.
Nucleic Acids Res., 37, 2009
|
|
2LSX
 
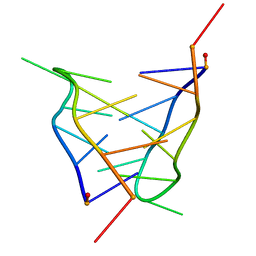 | | Solution structure of a mini i-motif | | Descriptor: | DNA (5'-D(P*TP*CP*GP*TP*TP*TP*CP*GP*TP*T)-3') | | Authors: | Escaja, N, Viladoms, J, Garavis, M, Villasante, A, Pedroso, E, Gonzalez, C. | | Deposit date: | 2012-05-09 | | Release date: | 2012-10-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A minimal i-motif stabilized by minor groove G:T:G:T tetrads.
Nucleic Acids Res., 40, 2012
|
|
2LE2
 
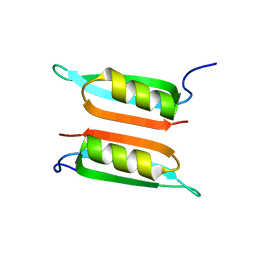 | | Novel dimeric structure of phage phi29-encoded protein p56: Insights into Uracil-DNA glycosylase inhibition | | Descriptor: | P56 | | Authors: | Asensio, J, Perez-Lago, L, Lazaro, J.M, Gonzalez, C, Serrano-Heras, G, Salas, M. | | Deposit date: | 2011-06-06 | | Release date: | 2011-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Novel dimeric structure of phage 29-encoded protein p56: insights into uracil-DNA glycosylase inhibition.
Nucleic Acids Res., 39, 2011
|
|
5NIP
 
 | |
5OGA
 
 | | Structure of minimal i-motif domain | | Descriptor: | DNA (5'-D(*TP*(DCP)P*GP*TP*TP*CP*(DCP)P*GP*TP*TP*TP*TP*TP*CP*GP*TP*TP*CP*CP*GP*T)-3') | | Authors: | Mir, B, Serrano, I, Buitrago, D, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2017-07-12 | | Release date: | 2017-11-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Prevalent Sequences in the Human Genome Can Form Mini i-Motif Structures at Physiological pH.
J. Am. Chem. Soc., 139, 2017
|
|
5MJX
 
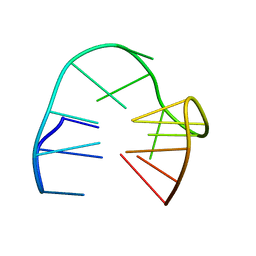 | | 2'F-ANA/DNA Chimeric TBA Quadruplex structure | | Descriptor: | DNA (5'-D(*GP*GP*(FT)P*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Lietard, J, Abou Assi, H, Gomez-Pinto, I, Gonzalez, C, Somoza, M.M, Damha, M.J. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Mapping the affinity landscape of Thrombin-binding aptamers on 2 F-ANA/DNA chimeric G-Quadruplex microarrays.
Nucleic Acids Res., 45, 2017
|
|
2N9H
 
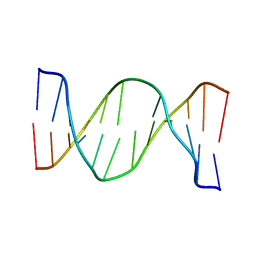 | | Glucose as a nuclease mimic in DNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*CP*(GL6)P*GP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*CP*TP*GP*CP*TP*AP*G)-3') | | Authors: | Gomez-Pinto, I, Vengut-Climent, E, Lucas, R, Avino, A, Eritja, R, Gonzalez-Ibanez, C, Morales, J, Muro, A, Penalver, P, Fonseca-Guerra, C, Bickelhaupt, M. | | Deposit date: | 2015-11-25 | | Release date: | 2016-08-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Glucose-Nucleobase Pseudo Base Pairs: Biomolecular Interactions within DNA.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2N9F
 
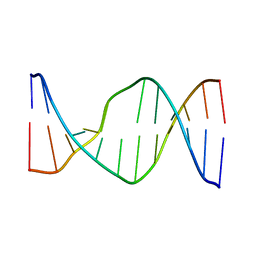 | | Glucose as non natural nucleobase | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*CP*GP*GP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*CP*(4JA)P*GP*CP*TP*AP*G)-3') | | Authors: | Gomez-Pinto, I, Vengut-Climent, E, Lucas, R, Avino, A, Eritja, R, Gonzalez-Ibanez, C, Morales, J, Penalver, P, Fonseca-Guerra, C, Bickelhaupt, M. | | Deposit date: | 2015-11-20 | | Release date: | 2016-08-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Glucose-Nucleobase Pseudo Base Pairs: Biomolecular Interactions within DNA.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2LYG
 
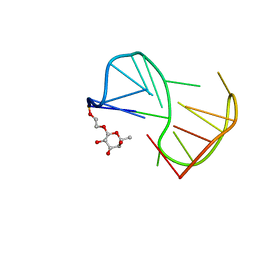 | | Fuc_TBA | | Descriptor: | 2-hydroxyethyl 6-deoxy-beta-L-galactopyranoside, DNA (5'-D(P*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Gomez-Pinto, I, Vengut-Climent, E, Lucas, R, Avio, A, Eritja, R, Gonzalez-Ibaez, C, Morales, J. | | Deposit date: | 2012-09-18 | | Release date: | 2014-01-29 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Carbohydrate-DNA interactions at G-quadruplexes: folding and stability changes by attaching sugars at the 5'-end.
Chemistry, 19, 2013
|
|
6Z7O
 
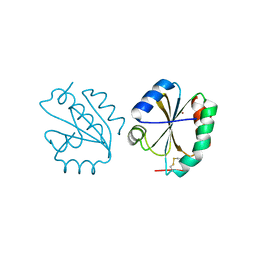 | | Crystal structure of Thioredoxin T from Drosophila melanogaster | | Descriptor: | Thioredoxin-T, ZINC ION | | Authors: | Freier, R, Aragon, E, Baginski, B, Pluta, R, Martin-Malpartida, P, Torner, C, Gonzaez, C, Macias, M. | | Deposit date: | 2020-05-31 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structures of the germline-specific Deadhead and thioredoxin T proteins from Drosophila melanogaster reveal unique features among thioredoxins.
Iucrj, 8, 2021
|
|
8FNX
 
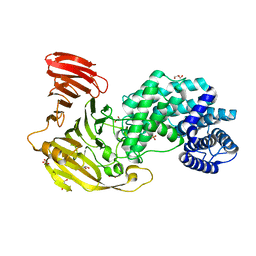 | | Crystal structure of Hyaluronate lyase B from Cutibacterium acnes | | Descriptor: | GLYCEROL, Hyaluronate lyase, PHOSPHATE ION | | Authors: | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | Deposit date: | 2022-12-28 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
8FYG
 
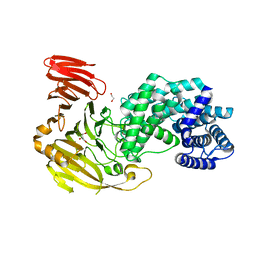 | | Crystal structure of Hyaluronate lyase A from Cutibacterium acnes | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hyaluronate lyase | | Authors: | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | Deposit date: | 2023-01-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
8G0O
 
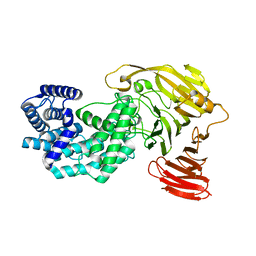 | | Crystal structure of Y281F mutant of Hyaluronate lyase B from Cutibacterium acnes | | Descriptor: | Hyaluronate lyase | | Authors: | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
6ZMU
 
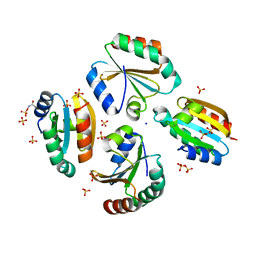 | | Crystal structure of the germline-specific thioredoxin protein Deadhead (Thioredoxin-1) from Drospohila melanogaster, P43212 | | Descriptor: | SODIUM ION, SULFATE ION, Thioredoxin-1 | | Authors: | Baginski, B, Pluta, R, Macias, M.J. | | Deposit date: | 2020-07-03 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the germline-specific Deadhead and thioredoxin T proteins from Drosophila melanogaster reveal unique features among thioredoxins.
Iucrj, 8, 2021
|
|
