7JIE
 
 | |
7RSW
 
 | | Crystal structure of group B human rotavirus VP8* | | Descriptor: | Outer capsid protein VP4, peptide | | Authors: | Hu, L, Salmen, W, Sankaran, B, Prasad, B.V. | | Deposit date: | 2021-08-11 | | Release date: | 2022-07-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Novel fold of rotavirus glycan-binding domain predicted by AlphaFold2 and determined by X-ray crystallography.
Commun Biol, 5, 2022
|
|
5KW9
 
 | |
4P26
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with A-type 2 HBGA | | Descriptor: | P domain of VP1, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-01 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P1V
 
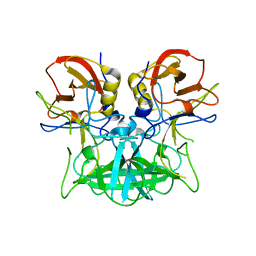 | | Structure of the P domain from a GI.7 Norovirus variant in complex with H-type 2 HBGA | | Descriptor: | P domain of VPI, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-02-27 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5497 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P2N
 
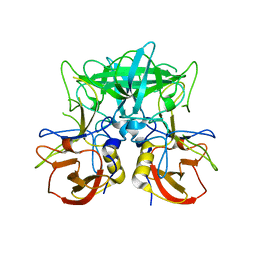 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeX HBGA | | Descriptor: | Major capsid protein, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P3I
 
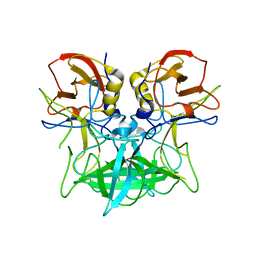 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeA HBGA. | | Descriptor: | P domain of VP1, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6941 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P25
 
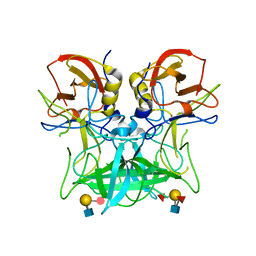 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeY HBGA. | | Descriptor: | Major capsid protein, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-01 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4991 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P12
 
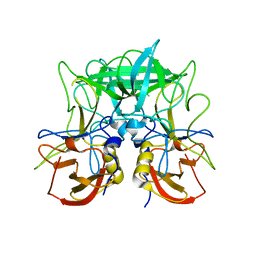 | | Native Structure of the P domain from a GI.7 Norovirus variant. | | Descriptor: | Major capsid protein | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-02-24 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6011 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
2GH8
 
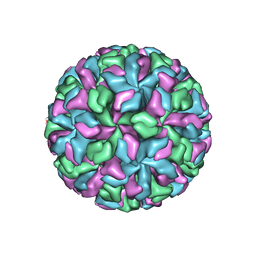 | | X-ray structure of a native calicivirus | | Descriptor: | Capsid protein | | Authors: | Chen, R. | | Deposit date: | 2006-03-26 | | Release date: | 2006-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structure of a native calicivirus: Structural insights into antigenic diversity and host specificity.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6AUK
 
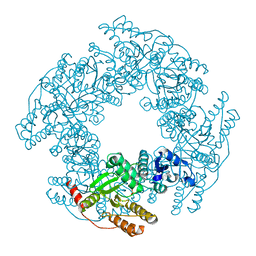 | |
6NIR
 
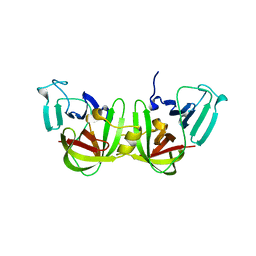 | | Crystal structure of a GII.4 norovirus HOV protease | | Descriptor: | HOV protease, HOV protease fragment | | Authors: | Prasad, B.V.V, Hu, L. | | Deposit date: | 2018-12-31 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | GII.4 Norovirus Protease Shows pH-Sensitive Proteolysis with a Unique Arg-His Pairing in the Catalytic Site.
J. Virol., 93, 2019
|
|
6O3V
 
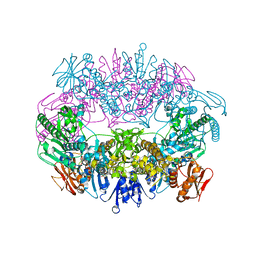 | | Crystal structure for RVA-VP3 | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-MONOPHOSPHATE, Protein VP3, ... | | Authors: | Kumar, D, Yu, X, Wang, Z, Hu, L, Prasad, V. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 2.7 angstrom cryo-EM structure of rotavirus core protein VP3, a unique capping machine with a helicase activity.
Sci Adv, 6, 2020
|
|
5VX5
 
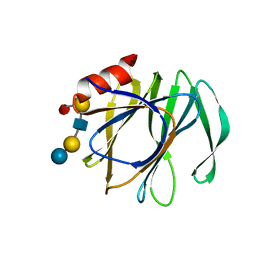 | | VP8* of a G2P[4] Human Rotavirus in complex with LNFP1 | | Descriptor: | Outer capsid protein VP4, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hu, L, Venkataram Prasad, B.V. | | Deposit date: | 2017-05-23 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.285 Å) | | Cite: | Glycan recognition in globally dominant human rotaviruses.
Nat Commun, 9, 2018
|
|
5VX4
 
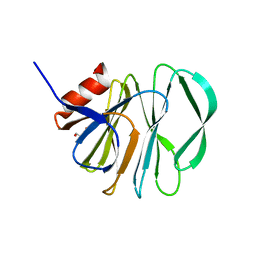 | | VP8* of a G2P[4] Human Rotavirus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Outer capsid protein VP4 | | Authors: | Hu, L, Venkataram Prasad, B.V. | | Deposit date: | 2017-05-23 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Glycan recognition in globally dominant human rotaviruses.
Nat Commun, 9, 2018
|
|
5VX8
 
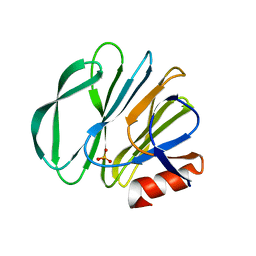 | | VP8* of P[6] Human Rotavirus RV3 | | Descriptor: | Outer capsid protein VP4, SULFATE ION | | Authors: | Hu, L, Venkataram Prasad, B.V. | | Deposit date: | 2017-05-23 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Glycan recognition in globally dominant human rotaviruses.
Nat Commun, 9, 2018
|
|
4YG0
 
 | | Structural basis of glycan recognition in neonate-specific rotaviruses | | Descriptor: | Outer capsid protein VP4, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hu, L, Prasad, B.V.V. | | Deposit date: | 2015-02-25 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.285 Å) | | Cite: | Structural basis of glycan specificity in neonate-specific bovine-human reassortant rotavirus.
Nat Commun, 6, 2015
|
|
4YG3
 
 | |
4YFW
 
 | |
4YG6
 
 | | Structural basis of glycan recognition in neonate-specific rotaviruses | | Descriptor: | Outer capsid protein VP8*, PHOSPHATE ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, L, Prasad, B.V.V. | | Deposit date: | 2015-02-25 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural basis of glycan specificity in neonate-specific bovine-human reassortant rotavirus.
Nat Commun, 6, 2015
|
|
4YFZ
 
 | | Structural basis of glycan recognition in neonate-specific rotaviruses | | Descriptor: | Outer capsid protein VP4, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hu, L, Prasad, B.V.V. | | Deposit date: | 2015-02-25 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of glycan specificity in neonate-specific bovine-human reassortant rotavirus.
Nat Commun, 6, 2015
|
|
4G0A
 
 | |
4G0J
 
 | |
