3DP5
 
 | | Crystal structure of Geobacter sulfurreducens OmcF with N-terminal Strep-tag II | | Descriptor: | Cytochrome c family protein, HEME C, SULFATE ION | | Authors: | Lukat, P, Hoffmann, M, Einsle, O. | | Deposit date: | 2008-07-07 | | Release date: | 2008-09-02 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal packing of the c(6)-type cytochrome OmcF from Geobacter sulfurreducens is mediated by an N-terminal Strep-tag II
ACTA CRYSTALLOGR.,SECT.D, 64, 2008
|
|
4L4Q
 
 | | Methionine Adenosyltransferase | | Descriptor: | S-adenosylmethionine synthase | | Authors: | Schlesier, J, Siegrist, J, Gerhardt, S, Andexer, J.N, Einsle, O. | | Deposit date: | 2013-06-09 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional characterisation of the methionine adenosyltransferase from Thermococcus kodakarensis.
Bmc Struct.Biol., 13, 2013
|
|
6Y77
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H326A | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DINUCLEAR COPPER ION, FORMIC ACID, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
6Y72
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H178A | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
6Y71
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H130A | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
6Y6Y
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H129A | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
6Y7E
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H494A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
6Y7D
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H433A | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
4LYS
 
 | | Crystal Structure of BRD4(1) bound to Colchiceine | | Descriptor: | Bromodomain-containing protein 4, N-[(7S)-10-hydroxy-1,2,3-trimethoxy-9-oxo-5,6,7,9-tetrahydrobenzo[a]heptalen-7-yl]acetamide, SODIUM ION | | Authors: | Wohlwend, D, Gerhardt, S, Einsle, O. | | Deposit date: | 2013-07-31 | | Release date: | 2014-01-15 | | Last modified: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 4-Acyl pyrroles: mimicking acetylated lysines in histone code reading.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4LYW
 
 | | Crystal Structure of BRD4(1) bound to inhibitor XD14 | | Descriptor: | 4-acetyl-N-[5-(diethylsulfamoyl)-2-hydroxyphenyl]-3-ethyl-5-methyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Wohlwend, D, Gerhardt, S, Einsle, O. | | Deposit date: | 2013-07-31 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-Acyl pyrroles: mimicking acetylated lysines in histone code reading.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4LZR
 
 | | Crystal Structure of BRD4(1) bound to Colchicine | | Descriptor: | Bromodomain-containing protein 4, N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide | | Authors: | Wohlwend, D, Gerhardt, S, Einsle, O, Huegle, M. | | Deposit date: | 2013-08-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 4-Acyl pyrroles: mimicking acetylated lysines in histone code reading.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4LZS
 
 | | Crystal Structure of BRD4(1) bound to inhibitor XD46 | | Descriptor: | 4-acetyl-3-ethyl-N,5-dimethyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Wohlwend, D, Huegle, M, Einsle, O, Gerhardt, S. | | Deposit date: | 2013-08-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 4-Acyl pyrroles: mimicking acetylated lysines in histone code reading.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
6ZMQ
 
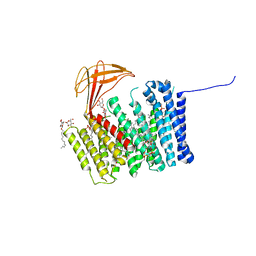 | | Cytochrome c Heme Lyase CcmF | | Descriptor: | Cytochrome C-type biogenesis protein ccmF, DODECYL-BETA-D-MALTOSIDE, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Brausemann, A, Einsle, O. | | Deposit date: | 2020-07-03 | | Release date: | 2021-05-19 | | Last modified: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Architecture of the membrane-bound cytochrome c heme lyase CcmF.
Nat.Chem.Biol., 17, 2021
|
|
4OQZ
 
 | | Streptomyces aurantiacus imine reductase | | Descriptor: | Putative oxidoreductase YfjR | | Authors: | Schneider, L.K, Huber, T, Gerhardt, S, Muller, M, Einsle, O. | | Deposit date: | 2014-02-10 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Direct Reductive Amination of Ketones: Structure and Activity of S-Selective Imine Reductases from Streptomyces.
CHEMCATCHEM, 2014
|
|
4OQY
 
 | | Streptomyces sp. GF3546 imine reductase | | Descriptor: | (S)-imine reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Schneider, L.K, Huber, T, Gerhardt, S, Muller, M, Einsle, O. | | Deposit date: | 2014-02-10 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Direct Reductive Amination of Ketones: Structure and Activity of S-Selective Imine Reductases from Streptomyces.
CHEMCATCHEM, 2014
|
|
1L5H
 
 | | FeMo-cofactor Deficient Nitrogenase MoFe Protein | | Descriptor: | CALCIUM ION, FE(8)-S(7) CLUSTER, nitrogenase molybdenum-iron protein alpha chain, ... | | Authors: | Schmid, B, Ribbe, M.W, Einsle, O, Yoshida, M, Thomas, L.M, Dean, D.R, Rees, D.C, Burgess, B.K. | | Deposit date: | 2002-03-06 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a cofactor-deficient nitrogenase MoFe protein.
Science, 296, 2002
|
|
1M2A
 
 | | Crystal structure at 1.5 Angstroms resolution of the wild type thioredoxin-like [2Fe-2S] ferredoxin from Aquifex aeolicus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, SULFATE ION, ZINC ION, ... | | Authors: | Yeh, A.P, Ambroggio, X.I, Andrade, S.L.A, Einsle, O, Chatelet, C, Meyer, J, Rees, D.C. | | Deposit date: | 2002-06-22 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structures of the wild type and
Cys-55-->Ser and Cys-59-->Ser variants of the thioredoxin-like
[2Fe-2S] ferredoxin from Aquifex aeolicus
J.Biol.Chem., 277, 2002
|
|
1M1Y
 
 | | Chemical Crosslink of Nitrogenase MoFe Protein and Fe Protein | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Schmid, B, Einsle, O, Chiu, H.J, Willing, A, Yoshida, M, Howard, J.B, Rees, D.C. | | Deposit date: | 2002-06-20 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical and Structural Characterization of the Crosslinked Complex of Nitrogenase: Comparison to the ADP-AlF4- Stabilized Structure
Biochemistry, 41, 2002
|
|
1M2D
 
 | | Crystal structure at 1.05 Angstroms resolution of the Cys59Ser variant of the thioredoxin-like [2Fe-2S] ferredoxin from Aquifex aeolicus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, [2Fe-2S] ferredoxin | | Authors: | Yeh, A.P, Ambroggio, X.I, Andrade, S.L.A, Einsle, O, Chatelet, C, Meyer, J, Rees, D.C. | | Deposit date: | 2002-06-22 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | High-resolution crystal structures
of the wild type and Cys-55-->Ser and
Cys-59-->Ser variants of the thioredoxin-like
[2Fe-2S] ferredoxin from Aquifex aeolicus
J.Biol.Chem., 277, 2002
|
|
1M34
 
 | | Nitrogenase Complex From Azotobacter Vinelandii Stabilized By ADP-Tetrafluoroaluminate | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Schmid, B, Einsle, O, Chiu, H.-J, Willing, A, Yoshida, M, Howard, J.B, Rees, D.C. | | Deposit date: | 2002-06-27 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and Structural Characterization of the Crosslinked Complex of Nitrogenase: Comparison to the ADP-AlF4 Stabilized structure
Biochemistry, 41, 2002
|
|
1M2B
 
 | | Crystal structure at 1.25 Angstroms resolution of the Cys55Ser variant of the thioredoxin-like [2Fe-2S] ferredoxin from Aquifex aeolicus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, [2Fe-2S] ferredoxin | | Authors: | Yeh, A.P, Ambroggio, X.I, Andrade, S.L.A, Einsle, O, Chatelet, C, Meyer, J, Rees, D.C. | | Deposit date: | 2002-06-22 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High-resolution crystal structures
of the wild type and Cys-55-->Ser and
Cys-59-->Ser variants of the thioredoxin-like
[2Fe-2S] ferredoxin from Aquifex aeolicus
J.Biol.Chem., 277, 2002
|
|
4RIE
 
 | | Landomycin Glycosyltransferase LanGT2 | | Descriptor: | Glycosyl transferase homolog | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RIF
 
 | | Landomycin Glycosyltransferase LanGT2, carbasugar substrate complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-{[(R)-{[(1S,3R,4R,5S)-3,4-dihydroxy-5-methylcyclohexyl]oxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-3,4-dihydrothymidine, Glycosyl transferase homolog | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RIG
 
 | | Chimeric Glycosyltransferase LanGT2S8Ac | | Descriptor: | Glycosyl transferase, MAGNESIUM ION | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4RIH
 
 | | Chimeric Glycosyltransferase LanGT2S8Ac, carbasugar substrate complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-{[(R)-{[(1S,3R,4R,5S)-3,4-dihydroxy-5-methylcyclohexyl]oxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-3,4-dihydrothymidine, Glycosyl transferase homolog,Glycosyl transferase, MAGNESIUM ION | | Authors: | Tam, H.K, Gerhardt, S, Breit, B, Bechthold, A, Einsle, O. | | Deposit date: | 2014-10-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural Characterization of O- and C-Glycosylating Variants of the Landomycin Glycosyltransferase LanGT2.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
