6K8O
 
 | | Crystal structure of the Sulfolobus solfataricus topoisomerase III in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*AP*AP*GP*GP*TP*C)-3'), ZINC ION, topoisomerase III | | Authors: | Wang, H.Q, Zhang, J.H, Zheng, X, Zheng, Z.F, Dong, Y.H, Huang, L, Gong, Y. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the Sulfolobus solfataricus topoisomerase III reveal that its C-terminal novel zinc finger part is a unique decatenation domain
To Be Published
|
|
2G3F
 
 | | Crystal Structure of imidazolonepropionase complexed with imidazole-4-acetic acid sodium salt, a substrate homologue | | Descriptor: | 2H-IMIDAZOL-4-YLACETIC ACID, Imidazolonepropionase, ZINC ION | | Authors: | Yu, Y, Liang, Y.H, Su, X.D. | | Deposit date: | 2006-02-19 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A catalytic mechanism revealed by the crystal structures of the imidazolonepropionase from Bacillus subtilis
J.Biol.Chem., 281, 2006
|
|
2BB0
 
 | |
3BW4
 
 | |
3BW3
 
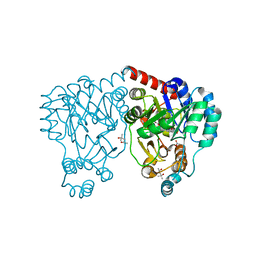 | | Crystal structures and site-directed mutagenesis study of nitroalkane oxidase from Streptomyces ansochromogenes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-nitropropane dioxygenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Li, Y.H, Gao, Z.Q, Hou, H.F. | | Deposit date: | 2008-01-08 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and site-directed mutagenesis of a nitroalkane oxidase from Streptomyces ansochromogenes
Biochem.Biophys.Res.Commun., 405, 2011
|
|
3BW2
 
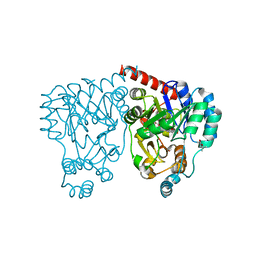 | | Crystal structures and site-directed mutagenesis study of nitroalkane oxidase from Streptomyces ansochromogenes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-nitropropane dioxygenase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, Y.H, Gao, Z.Q, Hou, H.F. | | Deposit date: | 2008-01-08 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and site-directed mutagenesis of a nitroalkane oxidase from Streptomyces ansochromogenes
Biochem.Biophys.Res.Commun., 405, 2011
|
|
4Q5W
 
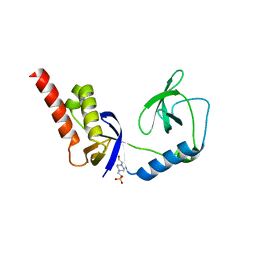 | | Crystal structure of extended-Tudor 9 of Drosophila melanogaster | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Maternal protein tudor | | Authors: | Ren, R, Liu, H, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | Deposit date: | 2014-04-17 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
4Q5Y
 
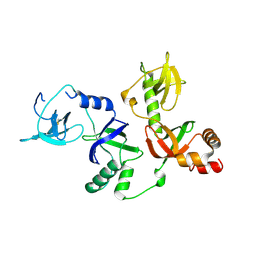 | | Crystal structure of extended-Tudor 10-11 of Drosophila melanogaster | | Descriptor: | Maternal protein tudor | | Authors: | Liu, H, Ren, R, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | Deposit date: | 2014-04-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
3Q5M
 
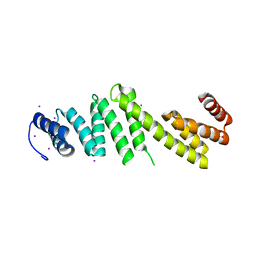 | | Crystal structure of Escherichia coli BamD | | Descriptor: | IODIDE ION, UPF0169 lipoprotein yfiO | | Authors: | Dong, C, Hou, H, Yang, X, Dong, Y, Shen, Y. | | Deposit date: | 2010-12-28 | | Release date: | 2011-12-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structure of Escherichia coli BamD and its functional implications in outer membrane protein assembly
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7FH9
 
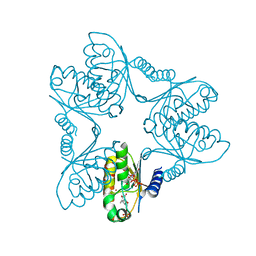 | |
7FH4
 
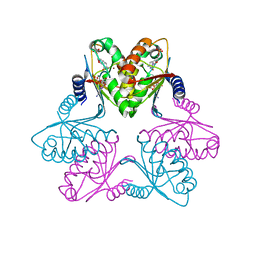 | | Chlorovirus PBCV-1 bi-functional dCMP/dCTP deaminase bi-DCD | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CMP/dCMP-type deaminase domain-containing protein, ... | | Authors: | She, Z. | | Deposit date: | 2021-07-29 | | Release date: | 2022-07-06 | | Last modified: | 2022-07-13 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Structural basis of a multi-functional deaminase in chlorovirus PBCV-1.
Arch.Biochem.Biophys., 727, 2022
|
|
3Q54
 
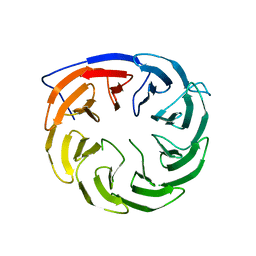 | | Crystal structure of Escherichia coli BamB | | Descriptor: | Outer membrane assembly lipoprotein YfgL | | Authors: | Dong, C, Hou, H, Yang, X. | | Deposit date: | 2010-12-27 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structure of Escherichia coli BamB and its interaction with POTRA domains of BamA.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4NSR
 
 | |
5C2O
 
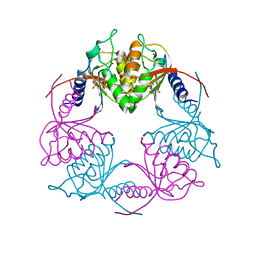 | | Crystal structure of Streptococcus mutans Deoxycytidylate Deaminase complexed with dTTP | | Descriptor: | MAGNESIUM ION, Putative deoxycytidylate deaminase, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Li, Y.H, Gao, Z.Q, Hou, H.F, Dong, Y.H. | | Deposit date: | 2015-06-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of the allosteric regulation of Streptococcus mutans 2'-deoxycytidylate deaminase.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4NSO
 
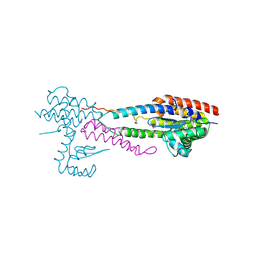 | | Crystal structure of the effector-immunity protein complex | | Descriptor: | Effector protein, Immunity protein | | Authors: | Dong, C. | | Deposit date: | 2013-11-28 | | Release date: | 2014-04-16 | | Last modified: | 2014-06-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for recognition of the type VI spike protein VgrG3 by a cognate immunity protein.
Febs Lett., 588, 2014
|
|
3TTM
 
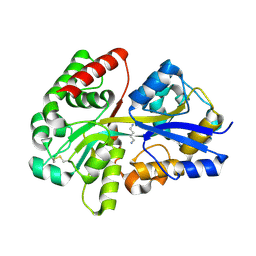 | | Crystal structure of SpuD in complex with putrescine | | Descriptor: | 1,4-DIAMINOBUTANE, Polyamine transport protein | | Authors: | Wu, D.H, Lim, S.C, Song, H.W. | | Deposit date: | 2011-09-15 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Substrate Binding Specificity Revealed by the Crystal Structures of Polyamine Receptors SpuD and SpuE from Pseudomonas aeruginosa
J.Mol.Biol., 416, 2012
|
|
3TTN
 
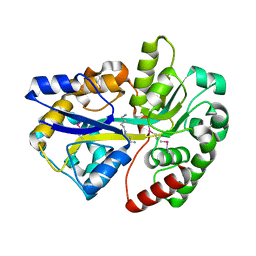 | |
3TTL
 
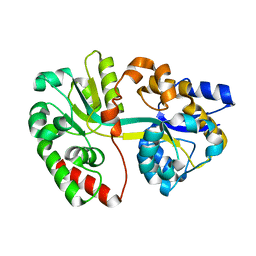 | | Crystal structure of apo-SpuE | | Descriptor: | Polyamine transport protein | | Authors: | Wu, D.H, Lim, S.C, Song, H.W. | | Deposit date: | 2011-09-14 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Substrate Binding Specificity Revealed by the Crystal Structures of Polyamine Receptors SpuD and SpuE from Pseudomonas aeruginosa
J.Mol.Biol., 416, 2012
|
|
3TTK
 
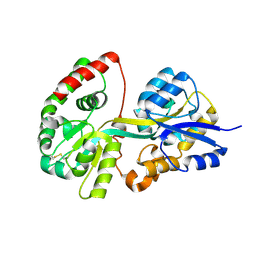 | | Crystal structure of apo-SpuD | | Descriptor: | Polyamine transport protein | | Authors: | Wu, D.H, Lim, S.C, Song, H.W. | | Deposit date: | 2011-09-14 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural Basis of Substrate Binding Specificity Revealed by the Crystal Structures of Polyamine Receptors SpuD and SpuE from Pseudomonas aeruginosa
J.Mol.Biol., 416, 2012
|
|
4X36
 
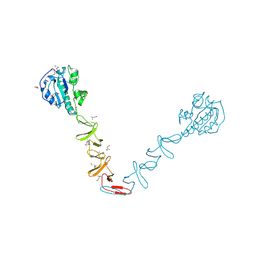 | | Crystal structure of the autolysin LytA from Streptococcus pneumoniae TIGR4 | | Descriptor: | Autolysin, CHOLINE ION, GLYCEROL, ... | | Authors: | Cheng, W, Li, Q, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2014-11-28 | | Release date: | 2015-05-27 | | Last modified: | 2015-06-24 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Full-length structure of the major autolysin LytA.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XO2
 
 | |
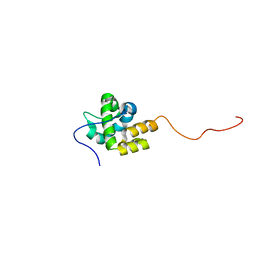
2MKZ
 
 | |
5YS1
 
 | | Crystal structure of Multicopper Oxidase CueO G304K mutant | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION | | Authors: | Wang, H.Q, Liu, X.Q, Zhao, J.T, Yue, Q.X, Yan, Y.H, Dong, Y.H, Fan, Y.L, Tian, J, Wu, N.F, Gong, Y. | | Deposit date: | 2017-11-12 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structures of multicopper oxidase CueO G304K mutant: structural basis of the increased laccase activity
Sci Rep, 8, 2018
|
|
5YS5
 
 | | Crystal structure of Multicopper Oxidase CueO G304K mutant with seven copper ions | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION | | Authors: | Wang, H.Q, Liu, X.Q, Zhao, J.T, Yue, Q.X, Yan, Y.H, Dong, Y.H, Fan, Y.L, Tian, J, Wu, N.F, Gong, Y. | | Deposit date: | 2017-11-13 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of multicopper oxidase CueO G304K mutant: structural basis of the increased laccase activity
Sci Rep, 8, 2018
|
|
3LCM
 
 | | Crystal structure of Smu.1420 from Streptococcus mutans UA159 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative oxidoreductase | | Authors: | Wang, Z.X, Su, X.-D. | | Deposit date: | 2010-01-11 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structural and biochemical characterization of MdaB from cariogenic Streptococcus mutans reveals an NADPH-specific quinone oxidoreductase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
