3J91
 
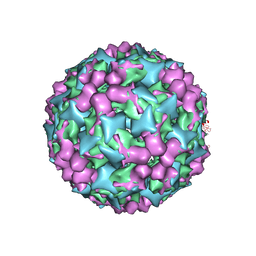 | | Cryo-electron microscopy of Enterovirus 71 (EV71) procapsid in complex with Fab fragments of neutralizing antibody 22A12 | | Descriptor: | VP0, VP1, VP3 | | Authors: | Shingler, K.L, Cifuente, J.O, Ashley, R.E, Makhov, A.M, Conway, J.F, Hafenstein, S. | | Deposit date: | 2014-11-24 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | The enterovirus 71 procapsid binds neutralizing antibodies and rescues virus infection in vitro.
J.Virol., 89, 2015
|
|
3JCX
 
 | | Canine Parvovirus complexed with Fab E | | Descriptor: | Capsid protein 2, Fab E heavy chain, Fab E light chain | | Authors: | Organtini, L.J, Iketani, S, Huang, K, Ashley, R.E, Makhov, A.M, Conway, J.F, Parrish, C.R, Hafenstein, S. | | Deposit date: | 2016-03-21 | | Release date: | 2016-07-20 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Near-Atomic Resolution Structure of a Highly Neutralizing Fab Bound to Canine Parvovirus.
J.Virol., 90, 2016
|
|
3JD7
 
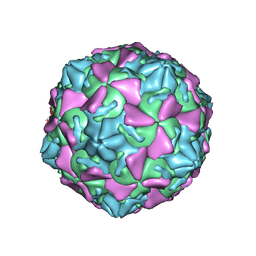 | | The novel asymmetric entry intermediate of a picornavirus captured with nanodiscs | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Lee, H, Shingler, K.L, Organtini, L.J, Ashley, R.E, Makhov, A.M, Conway, J.F, Hafenstein, S. | | Deposit date: | 2016-04-29 | | Release date: | 2016-09-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The novel asymmetric entry intermediate of a picornavirus captured with nanodiscs.
Sci Adv, 2, 2016
|
|
3J6L
 
 | | Kinetic and Structural Analysis of Coxsackievirus B3 Receptor Interactions and Formation of the A-particle | | Descriptor: | Coxsackievirus and adenovirus receptor, SULFATE ION | | Authors: | Organtini, L.J, Makhov, A.M, Conway, J.F, Hafenstein, S, Carson, S.D. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-09 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Kinetic and structural analysis of coxsackievirus b3 receptor interactions and formation of the a-particle.
J.Virol., 88, 2014
|
|
3J6N
 
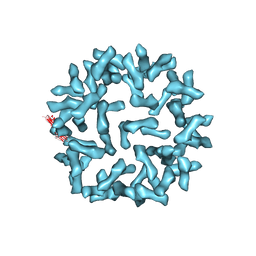 | | Kinetic and Structural Analysis of Coxsackievirus B3 Receptor Interactions and Formation of the A-particle | | Descriptor: | Coxsackie and adenovirus receptor | | Authors: | Organtini, L.J, Makhov, A.M, Conway, J.F, Hafenstein, S, Carson, S.D. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-09 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Kinetic and structural analysis of coxsackievirus b3 receptor interactions and formation of the a-particle.
J.Virol., 88, 2014
|
|
3J93
 
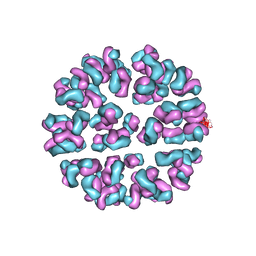 | | Fitting of Fab into the cryoEM density map of EV71 procapsid in complex with Fab22A12 | | Descriptor: | neutralizing antibody 22A12, heavy chain, light chain | | Authors: | Shingler, K.L, Cifuente, J.O, Ashley, R.E, Makhov, A.M, Conway, J.F, Hafenstein, S. | | Deposit date: | 2014-12-02 | | Release date: | 2014-12-24 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | The enterovirus 71 procapsid binds neutralizing antibodies and rescues virus infection in vitro.
J.Virol., 89, 2015
|
|
1C5E
 
 | | BACTERIOPHAGE LAMBDA HEAD PROTEIN D | | Descriptor: | GLYCEROL, HEAD DECORATION PROTEIN | | Authors: | Yang, F, Forrer, P, Dauter, Z, Pluckthun, A, Wlodawer, A. | | Deposit date: | 1999-11-18 | | Release date: | 2000-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD.
Nat.Struct.Biol., 7, 2000
|
|
5TJT
 
 | |
4E5Z
 
 | |
4E54
 
 | |
6Z6E
 
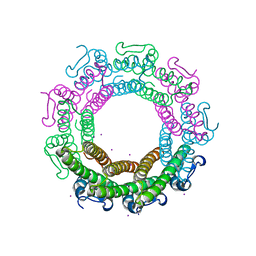 | | Crystal structure of the HK97 bacteriophage small terminase | | Descriptor: | IODIDE ION, Terminase small subunit | | Authors: | Fung, H.K.H, Chechik, M, Baumann, C.G, Antson, A.A. | | Deposit date: | 2020-05-28 | | Release date: | 2021-06-09 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of DNA packaging by a ring-type ATPase from an archetypal viral system.
Nucleic Acids Res., 50, 2022
|
|
6Z6D
 
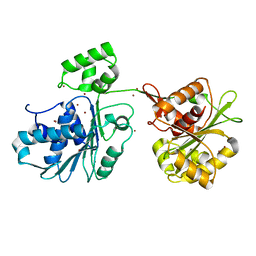 | | Crystal structure of the HK97 bacteriophage large terminase | | Descriptor: | BROMIDE ION, Terminase large subunit | | Authors: | Fung, H.K.H, Chechik, M, Baumann, C.G, Antson, A.A. | | Deposit date: | 2020-05-28 | | Release date: | 2021-06-09 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of DNA packaging by a ring-type ATPase from an archetypal viral system.
Nucleic Acids Res., 50, 2022
|
|
5LXK
 
 | | NMR structure of the C-terminal domain of the Bacteriophage T5 decoration protein pb10. | | Descriptor: | Decoration protein | | Authors: | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-08-02 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
5LXL
 
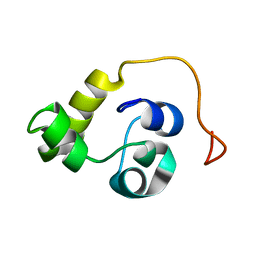 | | NMR structure of the N-terminal domain of the Bacteriophage T5 decoration protein pb10 | | Descriptor: | Decoration protein | | Authors: | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-04-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
7MEJ
 
 | |
7MDW
 
 | |
7ME7
 
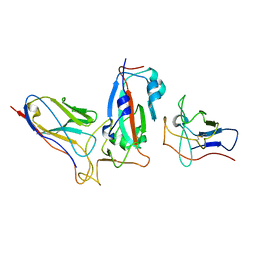 | |
7N9C
 
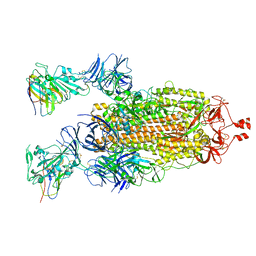 | |
7N9E
 
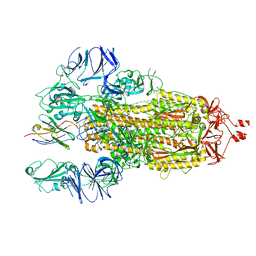 | |
7N9B
 
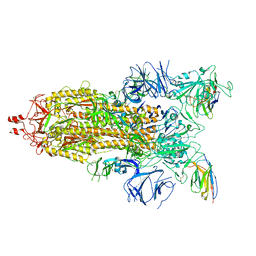 | |
7N9T
 
 | |
8CFA
 
 | |
8CEZ
 
 | |
8E16
 
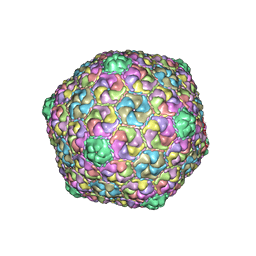 | | Mycobacterium phage Che8 | | Descriptor: | Major capsid protein, gp6 | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-08-09 | | Release date: | 2023-02-01 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8EC8
 
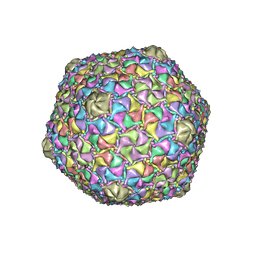 | | Mycobacterium phage Bobi | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-01 | | Release date: | 2023-02-01 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
