2A29
 
 | | The solution structure of the AMP-PNP bound nucleotide binding domain of KdpB | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Potassium-transporting ATPase B chain | | Authors: | Haupt, M, Bramkamp, M, Coles, M, Altendorf, K, Kessler, H. | | Deposit date: | 2005-06-22 | | Release date: | 2005-12-20 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The Holo-form of the Nucleotide Binding Domain of the KdpFABC Complex from Escherichia coli Reveals a New Binding Mode
J.Biol.Chem., 281, 2006
|
|
2A00
 
 | | The solution structure of the AMP-PNP bound nucleotide binding domain of KdpB | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Potassium-transporting ATPase B chain | | Authors: | Haupt, M, Bramkamp, M, Coles, M, Altendorf, K, Kessler, H. | | Deposit date: | 2005-06-15 | | Release date: | 2005-12-20 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The Holo-form of the Nucleotide Binding Domain of the KdpFABC Complex from Escherichia coli Reveals a New Binding Mode
J.Biol.Chem., 281, 2006
|
|
4C46
 
 | | ANDREI-N-LVPAS fused to GCN4 adaptors | | Descriptor: | BROMIDE ION, GENERAL CONTROL PROTEIN GCN4 | | Authors: | Albrecht, R, Alva, V, Ammelburg, M, Baer, K, Basina, E, Boichenko, I, Bonhoeffer, F, Braun, V, Chaubey, M, Chauhan, N, Chellamuthu, V.R, Coles, M, Deiss, S, Ewers, C.P, Forouzan, D, Fuchs, A, Groemping, Y, Hartmann, M.D, Hernandez Alvarez, B, Jeganantham, A, Kalev, I, Koenninger, U, Koiwai, K, Kopec, K.O, Korycinski, M, Laudenbach, B, Lehmann, K, Leo, J.C, Linke, D, Marialke, J, Martin, J, Mechelke, M, Michalik, M, Noll, A, Patzer, S.I, Scharfenberg, F, Schueckel, M, Shahid, S.A, Sulz, E, Ursinus, A, Wuertenberger, S, Zhu, H. | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Your Personalized Protein Structure: Andrei N. Lupas Fused to GCN4 Adaptors.
J.Struct.Biol., 186, 2014
|
|
1HZE
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF RIBOFLAVIN SYNTHASE FROM E. COLI | | Descriptor: | RIBOFLAVIN, RIBOFLAVIN SYNTHASE ALPHA CHAIN | | Authors: | Truffault, V, Coles, M, Diercks, T, Abelmann, K, Eberhardt, S, Luettgen, H, Bacher, A, Kessler, H. | | Deposit date: | 2001-01-24 | | Release date: | 2001-09-05 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of riboflavin synthase.
J.Mol.Biol., 309, 2001
|
|
1I18
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF RIBOFLAVIN SYNTHASE FROM E. COLI | | Descriptor: | RIBOFLAVIN, RIBOFLAVIN SYNTHASE ALPHA CHAIN | | Authors: | Truffault, V, Coles, M, Diercks, T, Abelmann, K, Eberhardt, S, Luettgen, H, Bacher, A, Kessler, H. | | Deposit date: | 2001-01-31 | | Release date: | 2001-09-05 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of riboflavin synthase.
J.Mol.Biol., 309, 2001
|
|
6H5H
 
 | |
4CQ4
 
 | | C-terminal fragment of Af1503-sol: transmembrane receptor Af1503 from Archaeoglobus fulgidus engineered for solubility | | Descriptor: | ENGINEERED VERSION OF TRANSMEMBRANE RECEPTOR AF1503 | | Authors: | Hartmann, M.D, Dunin-Horkawicz, S, Hulko, M, Martin, J, Coles, M, Lupas, A.N. | | Deposit date: | 2014-02-11 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Soluble Mutant of the Transmembrane Receptor Af1503 Features Strong Changes in Coiled-Coil Periodicity.
J.Struct.Biol., 186, 2014
|
|
2M3X
 
 | |
6FES
 
 | | Crystal structure of novel repeat protein BRIC2 fused to DARPin D12 | | Descriptor: | 1,2-ETHANEDIOL, D12_BRIC2, a synthetic protein,D12_BRIC2, ... | | Authors: | ElGamacy, M, Coles, M, Ernst, P, Zhu, H, Hartmann, M.D, Plueckthun, A, Lupas, A. | | Deposit date: | 2018-01-03 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | An Interface-Driven Design Strategy Yields a Novel, Corrugated Protein Architecture.
ACS Synth Biol, 7, 2018
|
|
6FF6
 
 | | Crystal structure of novel repeat protein BRIC1 | | Descriptor: | BRIC1 | | Authors: | ElGamacy, M, Coles, M, Ernst, P, Zhu, H, Hartmann, M.D, Plueckthun, A, Lupas, A.N. | | Deposit date: | 2018-01-03 | | Release date: | 2018-09-05 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Interface-Driven Design Strategy Yields a Novel, Corrugated Protein Architecture.
ACS Synth Biol, 7, 2018
|
|
4CTI
 
 | | Escherichia coli EnvZ histidine kinase catalytic part fused to Archaeoglobus fulgidus Af1503 HAMP domain | | Descriptor: | OSMOLARITY SENSOR PROTEIN ENVZ, AF1503 | | Authors: | Ferris, H.U, Coles, M, Lupas, A.N, Hartmann, M.D. | | Deposit date: | 2014-03-13 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | Crystallographic Snapshot of the Escherichia Coli Envz Histidine Kinase in an Active Conformation.
J.Struct.Biol., 186, 2014
|
|
2MVO
 
 | | Solution structure of the lantibiotic self-resistance lipoprotein MlbQ from Microbispora ATCC PTA-5024 | | Descriptor: | Putative lipoprotein | | Authors: | Pozzi, R, Schwartz, P, Linke, D, Kulik, A, Nega, M, Wohlleben, W, Stegmann, E, Coles, M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-07-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Distinct mechanisms contribute to immunity in the lantibiotic NAI-107 producer strain Microbispora ATCC PTA-5024.
Environ Microbiol, 18, 2016
|
|
2JV2
 
 | |
2KHM
 
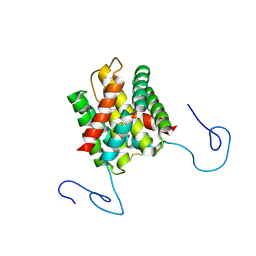 | | Structure of the C-terminal non-repetitive domain of the spider dragline silk protein ADF-3 | | Descriptor: | Fibroin-3 | | Authors: | Hagn, F.X, Eisoldt, L, Hardy, J.G, Vendrely, C, Coles, M, Scheibel, T, Kessler, H. | | Deposit date: | 2009-04-09 | | Release date: | 2010-04-14 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | A conserved spider silk domain acts as a molecular switch that controls fibre assembly
Nature, 465, 2010
|
|
2MUY
 
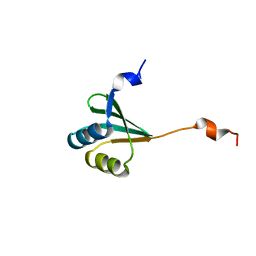 | | The solution structure of the FtsH periplasmic N-domain | | Descriptor: | ATP-dependent zinc metalloprotease FtsH | | Authors: | Scharfenberg, F, Serek-Heuberger, J, Martin, J, Lupas, A.N, Coles, M. | | Deposit date: | 2014-09-18 | | Release date: | 2015-01-28 | | Last modified: | 2015-02-25 | | Method: | SOLUTION NMR | | Cite: | Structure and Evolution of N-domains in AAA Metalloproteases.
J.Mol.Biol., 427, 2015
|
|
2MV3
 
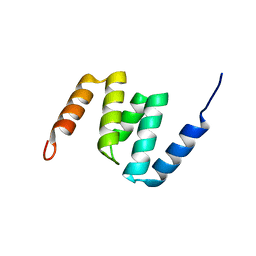 | | The N-domain of the AAA metalloproteinase Yme1 from Saccharomyces cerevisiae | | Descriptor: | Mitochondrial inner membrane i-AAA protease supercomplex subunit YME1 | | Authors: | Scharfenberg, F, Serek-Heuberger, J, Martin, J, Lupas, A.N, Coles, M. | | Deposit date: | 2014-09-22 | | Release date: | 2015-01-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and Evolution of N-domains in AAA Metalloproteases.
J.Mol.Biol., 427, 2015
|
|
2VXF
 
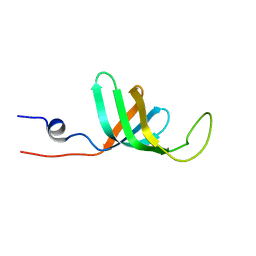 | |
2VXE
 
 | |
2RM4
 
 | |
4UZR
 
 | |
4V0B
 
 | | Escherichia coli FtsH hexameric N-domain | | Descriptor: | ATP-DEPENDENT ZINC METALLOPROTEASE FTSH, SULFATE ION | | Authors: | Serek-Heuberger, J, Martin, J, Lupas, A.N, Hartmann, M.D. | | Deposit date: | 2014-09-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and Evolution of N-Domains in Aaa Metalloproteases.
J.Mol.Biol., 427, 2015
|
|
1BY7
 
 | |
4V2Z
 
 | |
4V31
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Deoxyuridine | | Descriptor: | 2'-DEOXYURIDINE, CEREBLON ISOFORM 4, CITRATE ANION, ... | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2014-10-15 | | Release date: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thalidomide Mimics Uridine Binding to an Aromatic Cage in Cereblon.
J.Struct.Biol., 188, 2014
|
|
4V30
 
 | |
