1R6M
 
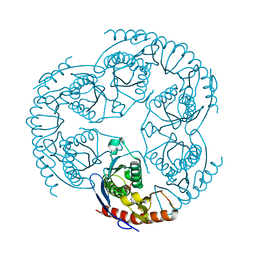 | | Crystal Structure Of The tRNA Processing Enzyme Rnase pH From Pseudomonas Aeruginosa In Complex With Phosphate | | Descriptor: | PHOSPHATE ION, Ribonuclease PH | | Authors: | Choi, J.M, Park, E.Y, Kim, J.H, Chang, S.K, Cho, Y. | | Deposit date: | 2003-10-15 | | Release date: | 2004-02-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the functional importance of the hexameric ring structure of RNase PH
J.BIOL.CHEM., 279, 2004
|
|
7CA3
 
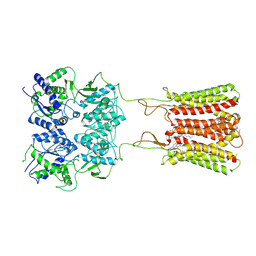 | | Cryo-EM structure of human GABA(B) receptor bound to the positive allosteric modulator rac-BHFF | | Descriptor: | (3S)-5,7-ditert-butyl-3-oxidanyl-3-(trifluoromethyl)-1-benzofuran-2-one, CHOLESTEROL, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Kim, Y, Jeong, E, Jeong, J, Kim, Y, Cho, Y. | | Deposit date: | 2020-06-08 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Basis for Activation of the Heterodimeric GABA B Receptor.
J.Mol.Biol., 432, 2020
|
|
3TAL
 
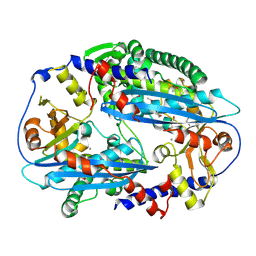 | | Crystal structure of NurA with manganese | | Descriptor: | DNA double-strand break repair protein nurA, GLYCEROL, MANGANESE (II) ION | | Authors: | Chae, J, Kim, Y.C, Cho, Y. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-23 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of the NurA-dAMP-Mn2+ complex
Nucleic Acids Res., 40, 2012
|
|
3TAZ
 
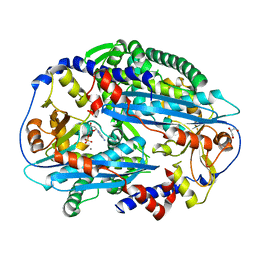 | | Crystal structure of NurA bound to dAMP and manganese | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, DNA double-strand break repair protein nurA, GLYCEROL, ... | | Authors: | Chae, J, Kim, Y.C, Cho, Y. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the NurA-dAMP-Mn2+ complex
Nucleic Acids Res., 40, 2012
|
|
7CA5
 
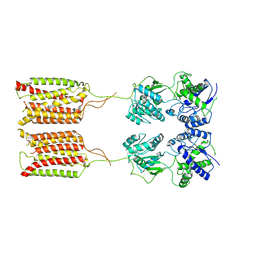 | | Cryo-EM structure of human GABA(B) receptor in apo state | | Descriptor: | Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2 | | Authors: | Kim, Y, Jeong, E, Jeong, J, Kim, Y, Cho, Y. | | Deposit date: | 2020-06-08 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural Basis for Activation of the Heterodimeric GABA B Receptor.
J.Mol.Biol., 432, 2020
|
|
2NWG
 
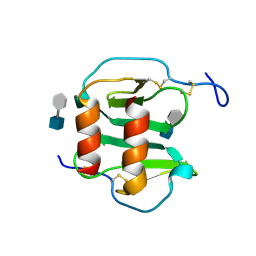 | | Structure of CXCL12:heparin disaccharide complex | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Stromal cell-derived factor 1 | | Authors: | Murphy, J.W, Cho, Y, Lolis, E. | | Deposit date: | 2006-11-14 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural and Functional Basis of CXCL12 (Stromal Cell-derived Factor-1{alpha}) Binding to Heparin
J.Biol.Chem., 282, 2007
|
|
3TAI
 
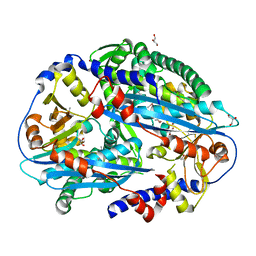 | | Crystal structure of NurA | | Descriptor: | DNA double-strand break repair protein nurA, GLYCEROL | | Authors: | Chae, J, Kim, Y.C, Cho, Y. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-23 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal structure of the NurA-dAMP-Mn2+ complex
Nucleic Acids Res., 40, 2012
|
|
7CUM
 
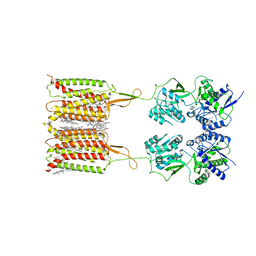 | | Cryo-EM structure of human GABA(B) receptor bound to the antagonist CGP54626 | | Descriptor: | (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid, CHOLESTEROL, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Kim, Y, Jeong, E, Jeong, J, Kim, Y, Cho, Y. | | Deposit date: | 2020-08-23 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural Basis for Activation of the Heterodimeric GABA B Receptor.
J.Mol.Biol., 432, 2020
|
|
6LHS
 
 | | High resolution structure of FANCA C-terminal domain (CTD) | | Descriptor: | Fanconi anemia complementation group A | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
6LHU
 
 | | High resolution structure of FANCA C-terminal domain (CTD) | | Descriptor: | Fanconi anemia complementation group A | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Kim, J, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
6LHW
 
 | | Structure of N-terminal and C-terminal domains of FANCA | | Descriptor: | Fanconi anemia complementation group A | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Kim, J, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.84 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
6LHV
 
 | | Structure of FANCA and FANCG Complex | | Descriptor: | Fanconi anemia complementation group A, Fanconi anemia complementation group G | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.59 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
2KLO
 
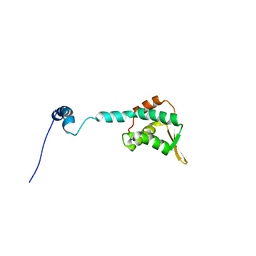 | | Structure of the Cdt1 C-terminal domain | | Descriptor: | DNA replication factor Cdt1 | | Authors: | Khayrutdinov, B.I, Bae, W.J, Yun, Y.M, Tsuyama, T, Kim, J.J, Hwang, E, Ryu, K.-S, Cheong, H.-K, Cheong, C, Karplus, P.A, Guntert, P, Tada, S, Jeon, Y.H, Cho, Y. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Cdt1 C-terminal domain: Conservation of the winged helix fold in replication licensing factors
Protein Sci., 18, 2009
|
|
2MJP
 
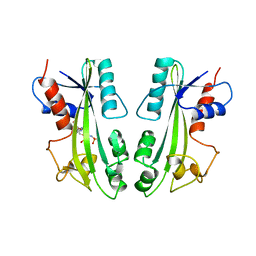 | | STRUCTURE-BASED IDENTIFICATION OF THE BIOCHEMICAL FUNCTION OF A HYPOTHETICAL PROTEIN FROM METHANOCOCCUS JANNASCHII:MJ0226 | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PYROPHOSPHATASE | | Authors: | Hwang, K.Y, Chung, J.H, Han, Y.S, Kim, S.H, Cho, Y, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 1999-01-27 | | Release date: | 2000-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based identification of a novel NTPase from Methanococcus jannaschii.
Nat.Struct.Biol., 6, 1999
|
|
6J10
 
 | | Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly | | Descriptor: | 6-cyclohexyl-4-methyl-1-oxidanyl-pyridin-2-one, Capsid protein | | Authors: | Park, S, Jin, M.S, Cho, Y, Kang, J, Kim, S, Park, M, Park, H, Kim, J, Park, S, Hwang, J, Kim, Y, Kim, Y.J. | | Deposit date: | 2018-12-27 | | Release date: | 2019-04-17 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly.
Nat Commun, 10, 2019
|
|
1M2K
 
 | | Sir2 homologue F159A mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2G
 
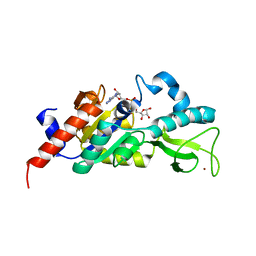 | | Sir2 homologue-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2J
 
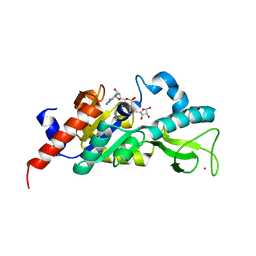 | | Sir2 homologue H80N mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2N
 
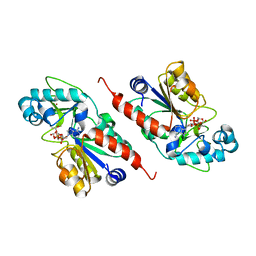 | |
1M2H
 
 | | Sir2 homologue S24A mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1N4M
 
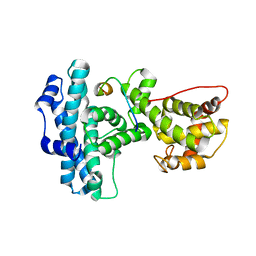 | | Structure of Rb tumor suppressor bound to the transactivation domain of E2F-2 | | Descriptor: | Retinoblastoma Pocket, Transcription factor E2F2 | | Authors: | Lee, C, Chang, J.H, Lee, H.S, Cho, Y. | | Deposit date: | 2002-10-31 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor
GENES DEV., 16, 2002
|
|
1N6A
 
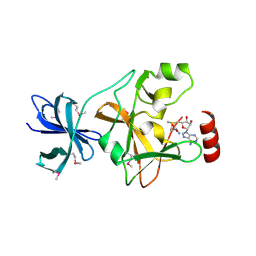 | | Structure of SET7/9 | | Descriptor: | S-ADENOSYLMETHIONINE, SET domain-containing protein 7 | | Authors: | Kwon, T.W, Chang, J.H, Kwak, E, Lee, C.W, Joachimiak, A, Kim, Y.C, Lee, J, Cho, Y. | | Deposit date: | 2002-11-09 | | Release date: | 2003-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet
EMBO J., 22, 2003
|
|
1N6C
 
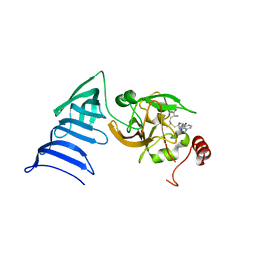 | | Structure of SET7/9 | | Descriptor: | S-ADENOSYLMETHIONINE, SET domain-containing protein 7 | | Authors: | Kwon, T.W, Chang, J.H, Cho, Y. | | Deposit date: | 2002-11-09 | | Release date: | 2003-02-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet
EMBO J., 22, 2003
|
|
5GOX
 
 | | Eukaryotic Rad50 Functions as A Rod-shaped Dimer | | Descriptor: | DNA repair protein RAD50, GLYCEROL, ZINC ION | | Authors: | Park, Y.B, Hohl, M, Padjasek, M, Jeong, E, Jin, K.S, Krezel, A, Petrini, J.H.J, Cho, Y. | | Deposit date: | 2016-07-30 | | Release date: | 2017-02-01 | | Last modified: | 2017-03-15 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Eukaryotic Rad50 functions as a rod-shaped dimer
Nat. Struct. Mol. Biol., 24, 2017
|
|
2EHO
 
 | | Crystal structure of human GINS complex | | Descriptor: | DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, GINS complex subunit 3, ... | | Authors: | Choi, J.M, Lim, H.S, Kim, J.J, Song, O.K, Cho, Y. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the human GINS complex
Genes Dev., 21, 2007
|
|
