6FW5
 
 | |
5LY8
 
 | | Structure of the CBM2 module of Lactobacillus casei BL23 phage J-1 evolved Dit. | | Descriptor: | MAGNESIUM ION, Tail component | | Authors: | Cambillau, C, Spinelli, S, Dieterle, M.-E, Piuri, M. | | Deposit date: | 2016-09-26 | | Release date: | 2017-03-22 | | Last modified: | 2017-05-24 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Evolved distal tail carbohydrate binding modules of Lactobacillus phage J-1: a novel type of anti-receptor widespread among lactic acid bacteria phages.
Mol. Microbiol., 104, 2017
|
|
5NGI
 
 | | Structure of XcpQN012 | | Descriptor: | Type II secretion system protein D | | Authors: | Cambillau, C, Roussel, A, Trinh, T.N. | | Deposit date: | 2017-03-17 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Unraveling the Self-Assembly of the Pseudomonas aeruginosa XcpQ Secretin Periplasmic Domain Provides New Molecular Insights into Type II Secretion System Secreton Architecture and Dynamics.
MBio, 8, 2017
|
|
4V5I
 
 | | Structure of the Phage P2 Baseplate in its Activated Conformation with Ca | | Descriptor: | CALCIUM ION, ORF15, ORF16, ... | | Authors: | Sciara, G, Bebeacua, C, Bron, P, Tremblay, D, Ortiz-Lombardia, M, Lichiere, J, van Heel, M, Campanacci, V, Moineau, S, Cambillau, C. | | Deposit date: | 2010-02-05 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (5.464 Å) | | Cite: | Structure of Lactococcal Phage P2 Baseplate and its Mechanism of Activation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1AGY
 
 | |
1C12
 
 | | INSIGHT IN ODORANT PERCEPTION: THE CRYSTAL STRUCTURE AND BINDING CHARACTERISTICS OF ANTIBODY FRAGMENTS DIRECTED AGAINST THE MUSK ODORANT TRASEOLIDE | | Descriptor: | PROTEIN (ANTIBODY FRAGMENT FAB), TRAZEOLIDE | | Authors: | Langedijk, A.C, Spinelli, S, Anguille, C, Hermans, P, Nederlof, J, Butenandt, J, Honegger, A, Cambillau, C, Pluckthun, A. | | Deposit date: | 1999-07-20 | | Release date: | 1999-08-14 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into odorant perception: the crystal structure and binding characteristics of antibody fragments directed against the musk odorant traseolide.
J.Mol.Biol., 292, 1999
|
|
4V96
 
 | | The structure of a 1.8 MDa viral genome injection device suggests alternative infection mechanisms | | Descriptor: | BPP, ORF46, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V, van Sinderen, D, Cambillau, C. | | Deposit date: | 2012-02-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1A3Y
 
 | |
1BU8
 
 | | RAT PANCREATIC LIPASE RELATED PROTEIN 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (PANCREATIC LIPASE RELATED PROTEIN 2) | | Authors: | Roussel, A, Cambillau, C. | | Deposit date: | 1998-09-14 | | Release date: | 1998-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and activity of rat pancreatic lipase-related protein 2.
J.Biol.Chem., 273, 1998
|
|
1BL9
 
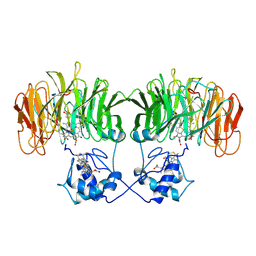 | | CONFORMATIONAL CHANGES OCCURRING UPON REDUCTION IN NITRITE REDUCTASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | HEME C, HEME D, HYDROXIDE ION, ... | | Authors: | Nurizzo, D, Cambillau, C, Tegoni, M. | | Deposit date: | 1998-07-20 | | Release date: | 1999-04-27 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Conformational changes occurring upon reduction and NO binding in nitrite reductase from Pseudomonas aeruginosa.
Biochemistry, 37, 1998
|
|
1B6D
 
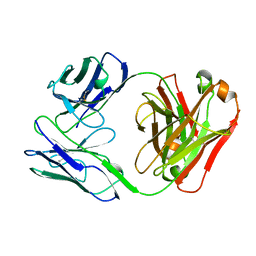 | | BENCE JONES PROTEIN DEL: AN ENTIRE IMMUNOGLOBULIN KAPPA LIGHT-CHAIN DIMER | | Descriptor: | IMMUNOGLOBULIN | | Authors: | Roussel, A, Spinelli, S, Deret, S, Aucouturier, P, Cambillau, C. | | Deposit date: | 1999-01-13 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The structure of an entire noncovalent immunoglobulin kappa light-chain dimer (Bence-Jones protein) reveals a weak and unusual constant domains association.
Eur.J.Biochem., 260, 1999
|
|
2PVS
 
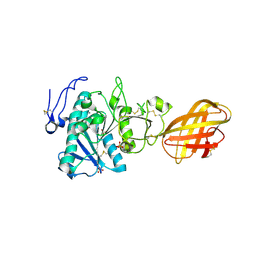 | | Structure of human pancreatic lipase related protein 2 mutant N336Q | | Descriptor: | CALCIUM ION, Pancreatic lipase-related protein 2, SULFATE ION | | Authors: | Spinelli, S, Eydoux, C, Carriere, F, Cambillau, C. | | Deposit date: | 2007-05-10 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human pancreatic lipase-related protein 2 with the lid in an open conformation.
Biochemistry, 47, 2008
|
|
6N38
 
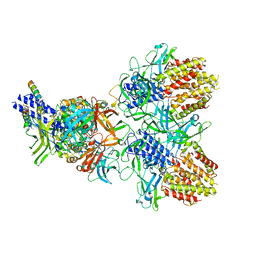 | | Structure of the type VI secretion system TssK-TssF-TssG baseplate subcomplex revealed by cryo-electron microscopy - full map sharpened | | Descriptor: | Putative type VI secretion protein, Unassigned protein | | Authors: | Park, Y.J, Lacourse, K.D, Cambillau, C, Seattle Structural Genomics Center for Infectious Disease (SSGCID), DiMaio, F, Mougous, J.D, Veesler, D. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the type VI secretion system TssK-TssF-TssG baseplate subcomplex revealed by cryo-electron microscopy.
Nat Commun, 9, 2018
|
|
5EFV
 
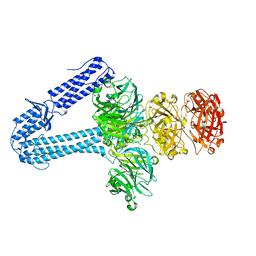 | | The host-recognition device of Staphylococcus aureus phage Phi11 | | Descriptor: | FE (III) ION, Phi ETA orf 56-like protein | | Authors: | Koc, C, Kuhner, P, Xia, G, Spinelli, S, Roussel, A, Cambillau, C, Stehle, T. | | Deposit date: | 2015-10-26 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the host-recognition device of Staphylococcus aureus phage 11.
Sci Rep, 6, 2016
|
|
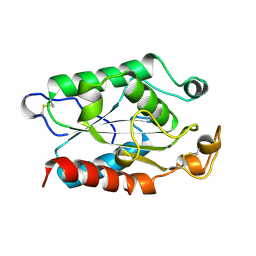
1XZA
 
 | |
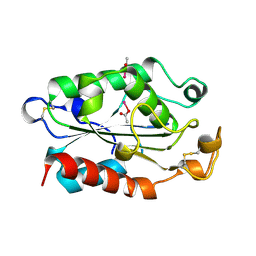
1XZB
 
 | |
4QGY
 
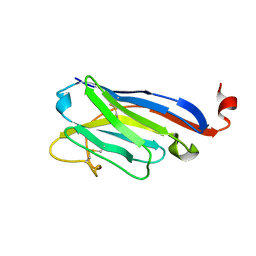 | | Camelid (llama) nanobody n25 (VHH) against type 6 secretion system TssM protein | | Descriptor: | nanobody n25, VH domain | | Authors: | Nguyen, V.S, Desmyter, A, Le, T.T.H, Durand, E, Kellenberger, C, Douzi, B, Spinelli, S, Cascales, E, Cambillau, C, Roussel, A. | | Deposit date: | 2014-05-26 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Inhibition of Type VI Secretion by an Anti-TssM Llama Nanobody.
Plos One, 10, 2015
|
|
4QLR
 
 | | Llama nanobody n02 raised against EAEC T6SS TssM | | Descriptor: | Llama nanobody n02 VH domain | | Authors: | Nguyen, V.S, Desmyter, A, Le, T.T.H, Durand, E, Kellenberger, C, Douzi, B, Spinelli, S, Cascales, E, Cambillau, C, Roussel, A. | | Deposit date: | 2014-06-13 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Type VI Secretion by an Anti-TssM Llama Nanobody.
Plos One, 10, 2015
|
|
4R90
 
 | | Anti CD70 Llama glama Fab 27B3 | | Descriptor: | Anti CD70 Llama glama Fab 27B3 Heavy chain, Anti CD70 Llama glama Fab 27B3 Light chain, CALCIUM ION, ... | | Authors: | Klarenbeek, A, El Mazouari, K, Desmyter, A, Blanchetot, C, Hultberg, A, Roovers, R.C, Cambillau, C, Spinelli, S, Del-Favero, J, Verrips, T, de Haard, H, Achour, I. | | Deposit date: | 2014-09-03 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Camelid Ig V genes reveal significant human homology not seen in therapeutic target genes, providing for a powerful therapeutic antibody platform.
MAbs, 7, 2015
|
|
4R96
 
 | | Structure of a Llama Glama Fab 48A2 against human cMet | | Descriptor: | Llama glama Fab 48A2 against human cMet H chain, Llama glama Fab 48A2 against human cMet L chain | | Authors: | Klarenbeek, A, El Mazouari, K, Desmyter, A, Blanchetot, C, Hultberg, A, Roovers, R.C, Cambillau, C, Spinelli, S, Del-Favero, J, Verrips, T, de Haard, H, Achour, I. | | Deposit date: | 2014-09-03 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Camelid Ig V genes reveal significant human homology not seen in therapeutic target genes, providing for a powerful therapeutic antibody platform.
MAbs, 7, 2015
|
|
1ZRU
 
 | | structure of the lactophage p2 receptor binding protein in complex with glycerol | | Descriptor: | GLYCEROL, lactophage p2 receptor binding protein | | Authors: | Spinelli, S, Tremblay, D.M, Tegoni, M, Blangy, S, Huyghe, C, Desmyter, A, Labrie, S, de Haard, H, Moineau, S, Cambillau, C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-05-22 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Receptor-binding protein of Lactococcus lactis phages: identification and characterization of the saccharide receptor-binding site.
J.Bacteriol., 188, 2006
|
|
4RGA
 
 | | Phage 1358 receptor binding protein in complex with the trisaccharide GlcNAc-Galf-GlcOMe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-galactofuranose-(1-6)-methyl alpha-D-glucopyranoside, Phage 1358 receptor binding protein (ORF20) | | Authors: | Spinelli, S, Mccabe, O, Farenc, C, Tremblay, D, Blangy, S, Oscarson, S, Moineau, S, Cambillau, C. | | Deposit date: | 2014-09-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The targeted recognition of Lactococcus lactis phages to their polysaccharide receptors.
Mol.Microbiol., 96, 2015
|
|
4ZS7
 
 | | Structural mimicry of receptor interaction by antagonistic IL-6 antibodies | | Descriptor: | Interleukin-6, Llama Fab fragment 68F2 heavy chain, Llama Fab fragment 68F2 light chain | | Authors: | Blanchetot, C, De Jonge, N, Desmyter, A, Ongenae, N, Hofman, E, Klarenbeek, A, Sadi, A, Hultberg, A, Kretz-Rommel, A, Spinelli, S, Loris, R, Cambillau, C, de Haard, H. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-04 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural Mimicry of Receptor Interaction by Antagonistic Interleukin-6 (IL-6) Antibodies.
J.Biol.Chem., 291, 2016
|
|
1XZJ
 
 | |
1XZC
 
 | |
