6ET1
 
 | |
6EYS
 
 | |
6ET2
 
 | |
6EYV
 
 | |
6ET0
 
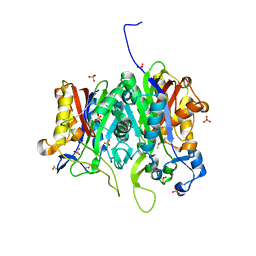 | | Crystal structure of PqsBC (C129A) mutant from Pseudomonas aeruginosa (crystal form 1) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Witzgall, F, Blankenfeldt, W. | | Deposit date: | 2017-10-25 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Alkylquinolone Repertoire of Pseudomonas aeruginosa is Linked to Structural Flexibility of the FabH-like 2-Heptyl-3-hydroxy-4(1H)-quinolone (PQS) Biosynthesis Enzyme PqsBC.
Chembiochem, 19, 2018
|
|
6ESZ
 
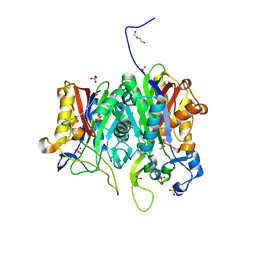 | |
6ET3
 
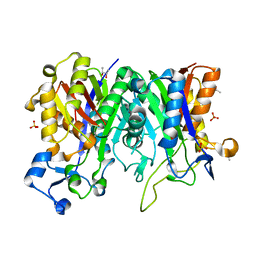 | | Crystal structure of PqsBC (C129S) mutant from Pseudomonas aeruginosa (crystal form 4) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, PqsB, PqsC, ... | | Authors: | Witzgall, F, Blankenfeldt, W. | | Deposit date: | 2017-10-25 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Alkylquinolone Repertoire of Pseudomonas aeruginosa is Linked to Structural Flexibility of the FabH-like 2-Heptyl-3-hydroxy-4(1H)-quinolone (PQS) Biosynthesis Enzyme PqsBC.
Chembiochem, 19, 2018
|
|
6EV2
 
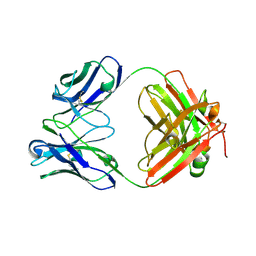 | | Crystal structure of antibody against schizophyllan in complex with laminarihexaose | | Descriptor: | Heavy chain, Light chain, beta-D-glucopyranose, ... | | Authors: | Sung, K.H, Josewski, J, Duebel, S, Blankenfeldt, W, Rau, U. | | Deposit date: | 2017-11-01 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Structural insights into antigen recognition of an anti-beta-(1,6)-beta-(1,3)-D-glucan antibody.
Sci Rep, 8, 2018
|
|
6FHP
 
 | | DAIP in complex with a C-terminal fragment of thermolysin | | Descriptor: | Dispase autolysis-inducing protein, Thermolysin | | Authors: | Schmelz, S, Fiebig, D, Fuchsbauer, H.L, Blankenfeldt, W, Scrima, A. | | Deposit date: | 2018-01-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Destructive twisting of neutral metalloproteases: the catalysis mechanism of the Dispase autolysis-inducing protein from Streptomyces mobaraensis DSM 40487.
FEBS J., 285, 2018
|
|
6FNT
 
 | |
6FRH
 
 | | Crystal structure of Ssp DnaB Mini-Intein variant M86 | | Descriptor: | Replicative DNA helicase,Replicative DNA helicase | | Authors: | Popp, M.A, Blankenfeldt, W, Gazdag, M.E, Matern, J.J.C, Mootz, H.D. | | Deposit date: | 2018-02-15 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A functional interplay between intein and extein sequences in protein splicing compensates for the essential block B histidine.
Chem Sci, 10, 2019
|
|
6FNQ
 
 | | Ergothioneine-biosynthetic methyltransferase EgtD in complex with N,N,N-trimethylhistidine (hercynine) | | Descriptor: | GLYCEROL, Histidine N-alpha-methyltransferase, MAGNESIUM ION, ... | | Authors: | Vit, A, Blankenfeldt, W, Seebeck, F.P. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inhibition and Regulation of the Ergothioneine Biosynthetic Methyltransferase EgtD.
ACS Chem. Biol., 13, 2018
|
|
6FNS
 
 | |
6FNR
 
 | | Ergothioneine-biosynthetic methyltransferase EgtD in complex with chlorohistidine | | Descriptor: | (2~{S})-2-chloranyl-3-(1~{H}-imidazol-5-yl)propanoic acid, GLYCEROL, Histidine N-alpha-methyltransferase, ... | | Authors: | Vit, A, Blankenfeldt, W, Seebeck, F.P. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Inhibition and Regulation of the Ergothioneine Biosynthetic Methyltransferase EgtD.
ACS Chem. Biol., 13, 2018
|
|
6FRE
 
 | | Crystal structure of G-1F/H73A mutant of Ssp DnaB Mini-Intein variant M86 | | Descriptor: | Replicative DNA helicase,Replicative DNA helicase | | Authors: | Popp, M.A, Blankenfeldt, W, Friedel, K, Mootz, H.D. | | Deposit date: | 2018-02-15 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | A functional interplay between intein and extein sequences in protein splicing compensates for the essential block B histidine.
Chem Sci, 10, 2019
|
|
6FRG
 
 | | Crystal structure of G-1F mutant of Ssp DnaB Mini-Intein variant M86 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Replicative DNA helicase, ... | | Authors: | Popp, M.A, Blankenfeldt, W, Friedel, K, Mootz, H.D. | | Deposit date: | 2018-02-15 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.535 Å) | | Cite: | A functional interplay between intein and extein sequences in protein splicing compensates for the essential block B histidine.
Chem Sci, 10, 2019
|
|
6GKX
 
 | |
6GKW
 
 | |
1SDJ
 
 | | X-RAY STRUCTURE OF YDDE_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET25. | | Descriptor: | Hypothetical protein yddE, SULFATE ION | | Authors: | Kuzin, A.P, Edstrom, W, Skarina, T, Korniyenko, Y, Savchenko, A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-02-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
5G3U
 
 | | The structure of the L-tryptophan oxidase VioA from Chromobacterium violaceum in complex with its inhibitor 2-(1H-indol-3-ylmethyl)prop-2- enoic acid | | Descriptor: | 2-[(1H-indol-3-yl)methyl]prop-2-enoic acid, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Krausze, J, Rabe, J, Moser, J. | | Deposit date: | 2016-05-01 | | Release date: | 2016-08-03 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.377 Å) | | Cite: | Biosynthesis of Violacein, Structure and Function of l-Tryptophan Oxidase VioA from Chromobacterium violaceum.
J.Biol.Chem., 291, 2016
|
|
1O7I
 
 | |
4AY7
 
 | | methyltransferase from Methanosarcina mazei | | Descriptor: | MAGNESIUM ION, METHYLCOBALAMIN: COENZYME M METHYLTRANSFERASE, ZINC ION | | Authors: | Hoeppner, A, Thomas, F, Rueppel, A, Hensel, R, Blankenfeld, W, Bayer, P, Faust, A. | | Deposit date: | 2012-06-18 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Corrinoid:Coenzyme M Methyltransferase Mtaa from Methanosarcina Mazei
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5G3T
 
 | | The structure of the L-tryptophan oxidase VioA from Chromobacterium violaceum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Krausze, J, Rabe, J, Moser, J. | | Deposit date: | 2016-05-01 | | Release date: | 2016-08-03 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biosynthesis of Violacein: Structure and Function of L-Tryptophan Oxidase Vioa Chromobacterium Violaceum
J.Biol.Chem., 291, 2016
|
|
5G3S
 
 | | The structure of the L-tryptophan oxidase VioA from Chromobacterium violaceum - Samarium derivative | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Krausze, J, Rabe, J, Moser, J. | | Deposit date: | 2016-05-01 | | Release date: | 2016-08-03 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (2.076 Å) | | Cite: | Biosynthesis of Violacein: Structure and Function of L-Tryptophan Oxidase Vioa Chromobacterium Violaceum
J.Biol.Chem., 291, 2016
|
|
6RMS
 
 | |
