5UUN
 
 | | Crystal structure of SARO_2595 from Novosphingobium aromaticivorans | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione S-transferase-like protein | | Authors: | Bingman, C.A, Kontur, W.S, Olmsted, C.N, Fox, B.G, Donohue, T.J. | | Deposit date: | 2017-02-17 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novosphingobium aromaticivoransuses a Nu-class glutathioneS-transferase as a glutathione lyase in breaking the beta-aryl ether bond of lignin.
J. Biol. Chem., 293, 2018
|
|
5UUO
 
 | | Crystal structure of SARO_2595 from Novosphingobium aromaticivorans | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase-like protein, ... | | Authors: | Bingman, C.A, Kontur, W.S, Olmsted, C.N, Fox, B.G, Donohue, T.J. | | Deposit date: | 2017-02-17 | | Release date: | 2018-02-28 | | Last modified: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Novosphingobium aromaticivoransuses a Nu-class glutathioneS-transferase as a glutathione lyase in breaking the beta-aryl ether bond of lignin.
J. Biol. Chem., 293, 2018
|
|
7LVZ
 
 | | Crystal structure of ADO | | Descriptor: | 2-aminoethanethiol dioxygenase, CHLORIDE ION, FE (II) ION, ... | | Authors: | Bingman, C.A, Fernandez, R.L, Smith, R.W, Fox, B.G, Brunold, T.C. | | Deposit date: | 2021-02-26 | | Release date: | 2022-01-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Crystal Structure of Cysteamine Dioxygenase Reveals the Origin of the Large Substrate Scope of This Vital Mammalian Enzyme.
Biochemistry, 60, 2021
|
|
4PED
 
 | | Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthes | | Descriptor: | Chaperone activity of bc1 complex-like, mitochondrial, SULFATE ION | | Authors: | Bingman, C.A, Smith, R, Joshi, S, Stefely, J.A, Reidenbach, A.G, Ulbrich, A, Oruganty, O, Floyd, B.J, Jochem, A, Saunders, J.M, Johnson, I.E, Wrobel, R.L, Barber, G.E, Lee, D, Li, S, Kannan, N, Coon, J.J, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mitochondrial ADCK3 Employs an Atypical Protein Kinase-like Fold to Enable Coenzyme Q Biosynthesis.
Mol.Cell, 57, 2015
|
|
7OBM
 
 | | Crystal structure of the human Prolyl Endopeptidase-Like protein short form (residues 90-727) | | Descriptor: | Prolyl endopeptidase-like | | Authors: | Rosier, K, McDevitt, M.T, Brendan, J.F, Marcaida, M.J, Bingman, C.A, Pagliarini, D.J, Creemers, J.W.M, Smith, R.W, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-10 | | Last modified: | 2022-02-02 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Prolyl endopeptidase-like is a (thio)esterase involved in mitochondrial respiratory chain function.
Iscience, 24, 2021
|
|
3TOS
 
 | | Crystal Structure of CalS11, Calicheamicin Methyltransferase | | Descriptor: | 1,2-ETHANEDIOL, CalS11, GLUTAMIC ACID, ... | | Authors: | Chang, A, Aceti, D.J, Beebe, E.T, Makino, S.-I, Wrobel, R.L, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-09-06 | | Release date: | 2011-10-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of CalS11, Calicheamicin methyltransferase
To be Published
|
|
6UI3
 
 | | GH5-4 broad specificity endoglucanase from Clostridum cellulovorans | | Descriptor: | 1,2-ETHANEDIOL, Cellulase | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
5F4Z
 
 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus | | Descriptor: | (1~{R},2~{R})-2,3-dihydro-1~{H}-indene-1,2-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, BABNIGG, G, BINGMAN, C.A, YENNAMALLI, R, LOHMAN, J, Chang, C.Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-12-03 | | Release date: | 2016-02-17 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus
To Be Published
|
|
7UZO
 
 | | Parathyroid hormone 1 receptor extracellular domain complexed with a peptide ligand containing one beta-amino acid | | Descriptor: | Parathyroid hormone/parathyroid hormone-related peptide receptor, Peptide from Parathyroid hormone-related protein, ZINC ION | | Authors: | Yu, Z, Bruchs, A.T, Bingman, C.A, Gellman, S.H. | | Deposit date: | 2022-05-09 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Altered signaling at the PTH receptor via modified agonist contacts with the extracellular domain provides a path to prolonged agonism in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UZP
 
 | | parathyroid hormone 1 receptor extracellular domain complexed with a peptide ligand containing three beta-amino acids | | Descriptor: | 1,2-ETHANEDIOL, PTHrP[1-36] 24,28,31 XCP, Parathyroid hormone/parathyroid hormone-related peptide receptor, ... | | Authors: | Yu, Z, Bruchs, A.T, Bingman, C.A, Gellman, S.H. | | Deposit date: | 2022-05-09 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Altered signaling at the PTH receptor via modified agonist contacts with the extracellular domain provides a path to prolonged agonism in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6Q1I
 
 | |
6PZ7
 
 | | GH5-4 broad specificity endoglucanase from Clostridium acetobutylicum | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Endoglucanase family 5 | | Authors: | Bianchetti, C.M, Bingman, C.A, Fox, B.G. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6WQV
 
 | | GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose | | Descriptor: | 1,2-ETHANEDIOL, Endoglucanase, NITRATE ION, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6WQP
 
 | | GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, Endoglucanase, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
1Q44
 
 | | Crystal Structure of an Arabidopsis Thaliana Putative Steroid Sulfotransferase | | Descriptor: | MALONIC ACID, Steroid Sulfotransferase | | Authors: | Phillips Jr, G.N, Smith, D.W, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of At2g03760, a putative steroid sulfotransferase from Arabidopsis thaliana
Proteins, 57, 2004
|
|
1Q45
 
 | | 12-0xo-phytodienoate reductase isoform 3 | | Descriptor: | 12-oxophytodienoate-10,11-reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Smith, D.W, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of Arabidopsis At2g06050, 12-oxophytodienoate reductase isoform 3
Proteins, 58, 2005
|
|
4FE3
 
 | |
4FZR
 
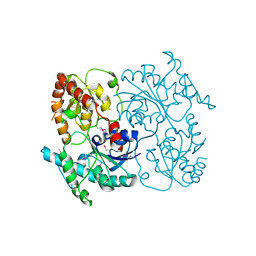 | | Crystal Structure of SsfS6, Streptomyces sp. SF2575 glycosyltransferase | | Descriptor: | SsfS6 | | Authors: | Wang, F, Zhou, M, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-07-07 | | Release date: | 2012-07-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Crystal structure of SsfS6, the putative C-glycosyltransferase involved in SF2575 biosynthesis.
Proteins, 81, 2013
|
|
4G2T
 
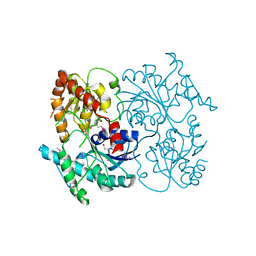 | | Crystal Structure of Streptomyces sp. SF2575 glycosyltransferase SsfS6, complexed with thymidine diphosphate | | Descriptor: | SsfS6, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Wang, F, Zhou, M, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-07-12 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Crystal structure of SsfS6, the putative C-glycosyltransferase involved in SF2575 biosynthesis.
Proteins, 81, 2013
|
|
6NIV
 
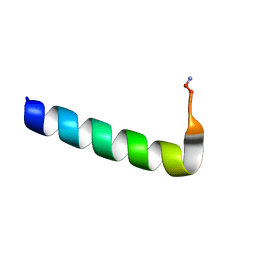 | | Racemic Phenol-Soluble Modulin Alpha 3 Peptide | | Descriptor: | Phenol-soluble modulin PSM-alpha-3 | | Authors: | Yao, Z, Cary, B.P, Bingman, C.A, Wang, C, Kreitler, D.F, Satyshur, K.A, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Use of a Stereochemical Strategy To Probe the Mechanism of Phenol-Soluble Modulin alpha 3 Toxicity.
J.Am.Chem.Soc., 141, 2019
|
|
6OWD
 
 | |
6O4M
 
 | | Racemic melittin | | Descriptor: | D-Melittin, Melittin, SULFATE ION | | Authors: | Kurgan, K.W, Bingman, C.A, Gellman, S.H, Forest, K.T. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Retention of Native Quaternary Structure in Racemic Melittin Crystals.
J.Am.Chem.Soc., 141, 2019
|
|
3BOM
 
 | | Crystal structure of trout hemoglobin at 1.35 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, Hemoglobin subunit alpha-4, Hemoglobin subunit beta-4, ... | | Authors: | Aranda IV, R, Bingman, C.A, Bitto, E, Wesenberg, G.E, Richards, M, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Trout Hemoglobin Crystal Structure at 1.35 Angstroms Resolution.
To be Published
|
|
3BVO
 
 | | Crystal structure of human co-chaperone protein HscB | | Descriptor: | Co-chaperone protein HscB, mitochondrial precursor, SULFATE ION, ... | | Authors: | Bitto, E, Bingman, C.A, McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-01-07 | | Release date: | 2008-01-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human J-type co-chaperone HscB reveals a tetracysteine metal-binding domain.
J.Biol.Chem., 283, 2008
|
|
3BUJ
 
 | | Crystal Structure of CalO2 | | Descriptor: | CalO2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | McCoy, J.G, Johnson, H.D, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2008-01-02 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural characterization of CalO2: a putative orsellinic acid P450 oxidase in the calicheamicin biosynthetic pathway.
Proteins, 74, 2009
|
|
