4EDK
 
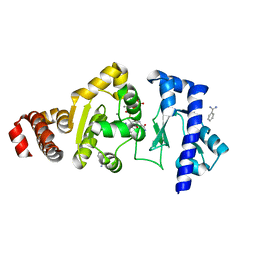 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to GTP and Manganese | | Descriptor: | BENZAMIDINE, DNA primase, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
5HE9
 
 | |
4FM9
 
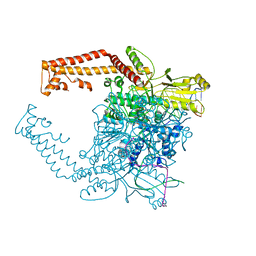 | | Human topoisomerase II alpha bound to DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), DNA topoisomerase 2-alpha, ... | | Authors: | Wendorff, T.J, Schmidt, B.H, Heslop, P, Austin, C.A, Berger, J.M. | | Deposit date: | 2012-06-15 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The Structure of DNA-Bound Human Topoisomerase II Alpha: Conformational Mechanisms for Coordinating Inter-Subunit Interactions with DNA Cleavage.
J.Mol.Biol., 424, 2012
|
|
5HE8
 
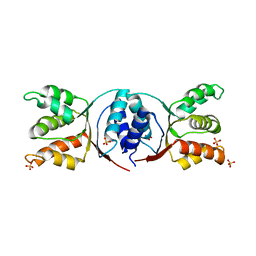 | | Bacterial initiation protein | | Descriptor: | Helicase loader, SULFATE ION | | Authors: | Hood, I.V, Berger, J.M. | | Deposit date: | 2016-01-05 | | Release date: | 2016-06-08 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bacterial initiation protein
To Be Published
|
|
4GFH
 
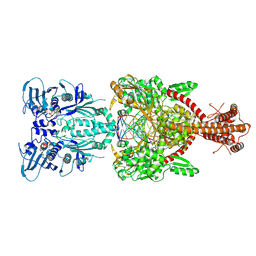 | | Topoisomerase II-DNA-AMPPNP complex | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*CP*GP*TP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*CP*GP*CP*GP*GP*TP*AP*GP*CP*AP*GP*TP*AP*GP*G)-3'), DNA (5'-D(P*CP*CP*TP*AP*CP*TP*GP*CP*TP*AP*C)-3'), ... | | Authors: | Schmidt, B.H, Osheroff, N, Berger, J.M. | | Deposit date: | 2012-08-03 | | Release date: | 2012-10-03 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (4.408 Å) | | Cite: | Structure of a topoisomerase II-DNA-nucleotide complex reveals a new control mechanism for ATPase activity.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5JJK
 
 | | Rho transcription termination factor bound to rA7 and 6 ADP-BeF3 molecules | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Thomsen, N.D, Lawson, M.R, Witkowsky, L.B, Qu, S, Berger, J.M. | | Deposit date: | 2016-04-24 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Molecular mechanisms of substrate-controlled ring dynamics and substepping in a nucleic acid-dependent hexameric motor.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5JJI
 
 | | Rho transcription termination factor bound to rU7 and 6 ADP-BeF3 molecules | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Thomsen, N.D, Lawson, M.R, Witkowsky, L.B, Qu, S, Berger, J.M. | | Deposit date: | 2016-04-24 | | Release date: | 2016-11-16 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Molecular mechanisms of substrate-controlled ring dynamics and substepping in a nucleic acid-dependent hexameric motor.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5JJL
 
 | | Rho transcription termination factor bound to rU8 and 5 ADP-BeF3 molecules | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Thomsen, N.D, Lawson, M.R, Witkowsky, L.B, Qu, S, Berger, J.M. | | Deposit date: | 2016-04-24 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular mechanisms of substrate-controlled ring dynamics and substepping in a nucleic acid-dependent hexameric motor.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
4XGC
 
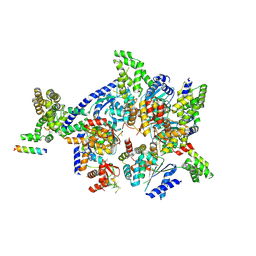 | | Crystal structure of the eukaryotic origin recognition complex | | Descriptor: | CHLORIDE ION, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | Authors: | Bleichert, F, Botchan, M.R, Berger, J.M. | | Deposit date: | 2014-12-30 | | Release date: | 2015-04-01 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the eukaryotic origin recognition complex.
Nature, 519, 2015
|
|
4Z98
 
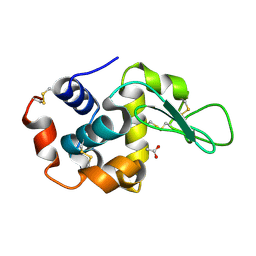 | | Crystal Structure of Hen Egg White Lysozyme using Serial X-ray Diffraction Data Collection | | Descriptor: | ACETATE ION, Lysozyme C | | Authors: | Murray, T.D, Lyubimov, A.Y, Ogata, C.M, Uervirojnangkoorn, M, Brunger, A.T, Berger, J.M. | | Deposit date: | 2015-04-10 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A high-transparency, micro-patternable chip for X-ray diffraction analysis of microcrystals under native growth conditions.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
8B4H
 
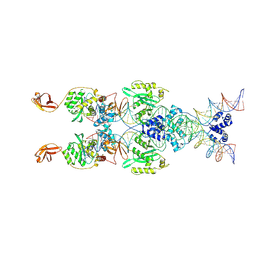 | | IstA transposase cleaved donor complex | | Descriptor: | DNA (55-MER) / right IS21 transposon end (insertion sequence IS5376), DNA (57-MER) / right IS21 transposon end (insertion sequence IS5376), MAGNESIUM ION, ... | | Authors: | Spinola-Amilibia, M, de la Gandara, A, Araujo-Bazan, L, Berger, J.M, Arias-Palomo, E. | | Deposit date: | 2022-09-20 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | IS21 family transposase cleaved donor complex traps two right-handed superhelical crossings.
Nat Commun, 14, 2023
|
|
1JL2
 
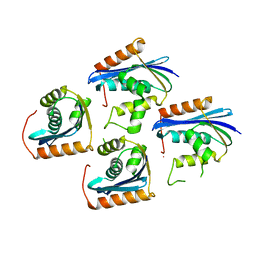 | |
1L8Q
 
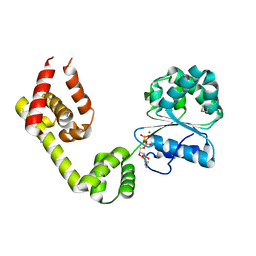 | |
437D
 
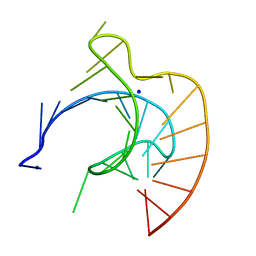 | | CRYSTAL STRUCTURE OF AN RNA PSEUDOKNOT FROM BEET WESTERN YELLOW VIRUS INVOLVED IN RIBOSOMAL FRAMESHIFTING | | Descriptor: | MAGNESIUM ION, RNA PSEUDOKNOT, SODIUM ION | | Authors: | Su, L, Chen, L, Egli, M, Berger, J.M, Rich, A. | | Deposit date: | 1998-11-24 | | Release date: | 1998-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot.
Nat.Struct.Biol., 6, 1999
|
|
3UC1
 
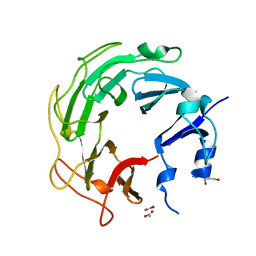 | | Mycobacterium tuberculosis gyrase type IIA topoisomerase C-terminal domain | | Descriptor: | ACETATE ION, CALCIUM ION, DNA gyrase subunit A, ... | | Authors: | Tretter, E.M, Berger, J.M. | | Deposit date: | 2011-10-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanisms for Defining Supercoiling Set Point of DNA Gyrase Orthologs: II. THE SHAPE OF THE GyrA SUBUNIT C-TERMINAL DOMAIN (CTD) IS NOT A SOLE DETERMINANT FOR CONTROLLING SUPERCOILING EFFICIENCY.
J.Biol.Chem., 287, 2012
|
|
4E2K
 
 | | The structure of the S. aureus DnaG RNA Polymerase Domain | | Descriptor: | BENZAMIDINE, DNA primase | | Authors: | Rymer, R.U, Solorio, F.A, Clement, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-08 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
4EDR
 
 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to UTP and Manganese | | Descriptor: | BENZAMIDINE, DNA primase, MANGANESE (II) ION, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
4EDT
 
 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to ppGpp and Manganese | | Descriptor: | BENZAMIDINE, DNA primase, GUANOSINE-5',3'-TETRAPHOSPHATE, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
4EDV
 
 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to pppGpp and Manganese | | Descriptor: | BENZAMIDINE, DNA primase, MANGANESE (II) ION, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
3C4O
 
 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) E73M/S130K/S146M complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Reynolds, K.A, Hanes, M.S, Thomson, J.M, Antczak, A.J, Berger, J.M, Bonomo, R.A, Kirsch, J.F, Handel, T.M. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational redesign of the SHV-1 beta-lactamase/beta-lactamase inhibitor protein interface.
J.Mol.Biol., 382, 2008
|
|
3C4P
 
 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) E73M complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Reynolds, K.A, Hanes, M.S, Thomson, J.M, Antczak, A.J, Berger, J.M, Bonomo, R.A, Kirsch, J.F, Handel, T.M. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Computational redesign of the SHV-1 beta-lactamase/beta-lactamase inhibitor protein interface.
J.Mol.Biol., 382, 2008
|
|
2HCB
 
 | | Structure of AMPPCP-bound DnaA from Aquifex aeolicus | | Descriptor: | Chromosomal replication initiator protein dnaA, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Erzberger, J.P, Mott, M.L, Berger, J.M. | | Deposit date: | 2006-06-15 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2HKJ
 
 | | Topoisomerase VI-B bound to radicicol | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, RADICICOL, ... | | Authors: | Corbett, K.D, Berger, J.M. | | Deposit date: | 2006-07-04 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for topoisomerase VI inhibition
by the anti-Hsp90 drug radicicol
Nucleic Acids Res., 34, 2006
|
|
2HT1
 
 | |
2IDC
 
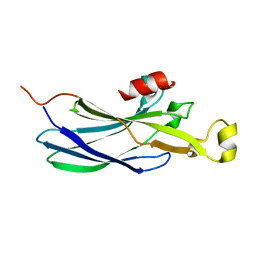 | | Structure of the Histone H3-Asf1 Chaperone Interaction | | Descriptor: | ANTI-SILENCING PROTEIN 1 AND HISTONE H3 CHIMERA | | Authors: | Antczak, A.J, Tsubota, T, Kaufman, P.D, Berger, J.M. | | Deposit date: | 2006-09-14 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the yeast histone H3-ASF1 interaction: implications for chaperone mechanism, species-specific interactions, and epigenetics.
Bmc Struct.Biol., 6, 2006
|
|
