6NP8
 
 | |
6NNP
 
 | |
6NNU
 
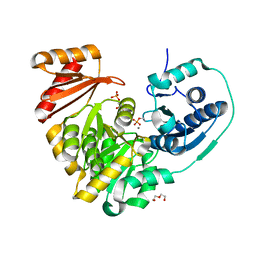 | | Xanthomonas citri Phospho-PGM in complex with glucose-1,6-phosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stiers, K.M, Beamer, L.J. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural and dynamical description of the enzymatic reaction of a phosphohexomutase.
Struct Dyn., 6, 2019
|
|
6NQH
 
 | |
6NNO
 
 | |
6NNS
 
 | |
6NPX
 
 | |
6NQG
 
 | |
6NNN
 
 | |
3PDK
 
 | | crystal structure of phosphoglucosamine mutase from B. anthracis | | Descriptor: | PHOSPHATE ION, Phosphoglucosamine mutase | | Authors: | Mehra-Chaudhary, R, Mick, J, Tanner, J.J, Henzl, M, Beamer, L.J. | | Deposit date: | 2010-10-22 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Bacillus anthracis Phosphoglucosamine Mutase, an Enzyme in the Peptidoglycan Biosynthetic Pathway.
J.Bacteriol., 193, 2011
|
|
3NA5
 
 | | Crystal structure of a bacterial phosphoglucomutase, an enzyme important in the virulence of several human pathogens. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Phosphoglucomutase | | Authors: | Mehra-Chaudhary, R, Beamer, L.J. | | Deposit date: | 2010-06-01 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bacterial phosphoglucomutase, an enzyme involved in the virulence of multiple human pathogens.
Proteins, 79, 2011
|
|
1K2Y
 
 | |
1K35
 
 | |
2FLU
 
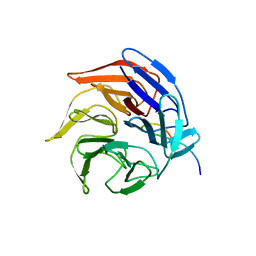 | | Crystal Structure of the Kelch-Neh2 Complex | | Descriptor: | Kelch-like ECH-associated protein 1, Nrf2 | | Authors: | Li, X, Lo, J, Beamer, L, Hannink, M. | | Deposit date: | 2006-01-06 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling.
Embo J., 25, 2006
|
|
6MNV
 
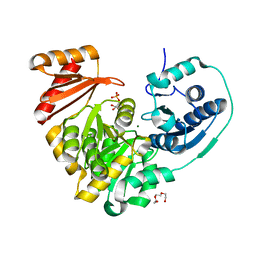 | | Crystal structure of X. citri phosphoglucomutase in complex with CH2FG1P | | Descriptor: | 1-deoxy-1-fluoro-2-O-phosphono-alpha-D-gluco-hept-2-ulopyranose, MAGNESIUM ION, Phosphomannomutase/phosphoglucomutase, ... | | Authors: | Beamer, L, Stiers, K. | | Deposit date: | 2018-10-03 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitory Evaluation of alpha PMM/PGM fromPseudomonas aeruginosa: Chemical Synthesis, Enzyme Kinetics, and Protein Crystallographic Study.
J.Org.Chem., 84, 2019
|
|
6MLF
 
 | |
6MLW
 
 | | Crystal structure of X. citri phosphoglucomutase in complex with 2-fluoro mannosyl-1-methyl-phosphonic acid | | Descriptor: | 2,6-anhydro-5,7-dideoxy-5-fluoro-7-phosphono-D-glycero-D-manno-heptitol, MAGNESIUM ION, Phosphoglucomutase | | Authors: | Beamer, L, Stiers, K. | | Deposit date: | 2018-09-28 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibitory Evaluation of alpha PMM/PGM fromPseudomonas aeruginosa: Chemical Synthesis, Enzyme Kinetics, and Protein Crystallographic Study.
J.Org.Chem., 84, 2019
|
|
6MLH
 
 | | Crystal structure of X. citri phosphoglucomutase in complex with GLUCOPYRANOSYL-1-METHYL-PHOSPHONIC ACID | | Descriptor: | (1S)-1,5-anhydro-1-(phosphonomethyl)-D-glucitol, MAGNESIUM ION, Phosphoglucomutase | | Authors: | Beamer, L, Stiers, K. | | Deposit date: | 2018-09-27 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibitory Evaluation of alpha PMM/PGM fromPseudomonas aeruginosa: Chemical Synthesis, Enzyme Kinetics, and Protein Crystallographic Study.
J.Org.Chem., 84, 2019
|
|
4MRQ
 
 | | Crystal Structure of wild-type unphosphorylated PMM/PGM | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L(+)-TARTARIC ACID, ... | | Authors: | Lee, Y, Beamer, L. | | Deposit date: | 2013-09-17 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Promotion of enzyme flexibility by dephosphorylation and coupling to the catalytic mechanism of a phosphohexomutase.
J.Biol.Chem., 289, 2014
|
|
4IL8
 
 | | Crystal structure of an H329A mutant of p. aeruginosa PMM/PGM | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphomannomutase/phosphoglucomutase | | Authors: | Lee, Y, Mehra-Chaudhary, R, Furdui, C, Beamer, L. | | Deposit date: | 2012-12-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of an essential active-site residue in the alpha-D-phosphohexomutase enzyme superfamily.
Febs J., 280, 2013
|
|
5UX5
 
 | | Structure of Proline Utilization A (PutA) from Corynebacterium freiburgense | | Descriptor: | BIFUNCTIONAL PROTEIN Proline utilization A (PutA), FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2017-02-22 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and characterization of a class 3B proline utilization A: Ligand-induced dimerization and importance of the C-terminal domain for catalysis.
J. Biol. Chem., 292, 2017
|
|
3RSM
 
 | | Crystal structure of S108C mutant of PMM/PGM | | Descriptor: | PHOSPHATE ION, Phosphomannomutase/phosphoglucomutase, ZINC ION | | Authors: | Akella, A, Anbanandam, A, Kelm, A, Wei, Y, Mehra-Chaudhary, R, Beamer, L, Van Doren, S. | | Deposit date: | 2011-05-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Solution NMR of a 463-residue phosphohexomutase: domain 4 mobility, substates, and phosphoryl transfer defect.
Biochemistry, 51, 2012
|
|
1LLI
 
 | | THE CRYSTAL STRUCTURE OF A MUTANT PROTEIN WITH ALTERED BUT IMPROVED HYDROPHOBIC CORE PACKING | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*CP*CP*AP*CP*TP*GP*GP*CP*GP*GP*TP*GP*A P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*TP*CP*AP*CP*CP*GP*CP*CP*AP*GP*TP*GP*G P*TP*AP*T)-3'), PROTEIN (LAMBDA REPRESSOR) | | Authors: | Lim, W.A, Hodel, A, Sauer, R.T, Richards, F.M. | | Deposit date: | 1994-03-25 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a mutant protein with altered but improved hydrophobic core packing.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
