1CZA
 
 | | MUTANT MONOMER OF RECOMBINANT HUMAN HEXOKINASE TYPE I COMPLEXED WITH GLUCOSE, GLUCOSE-6-PHOSPHATE, AND ADP | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, HEXOKINASE TYPE I, ... | | Authors: | Aleshin, A.E, Liu, X, Kirby, C, Bourenkov, G.P, Bartunik, H.D, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1999-09-01 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of mutant monomeric hexokinase I reveal multiple ADP binding sites and conformational changes relevant to allosteric regulation.
J.Mol.Biol., 296, 2000
|
|
1DGK
 
 | | MUTANT MONOMER OF RECOMBINANT HUMAN HEXOKINASE TYPE I WITH GLUCOSE AND ADP IN THE ACTIVE SITE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEXOKINASE TYPE I, PHOSPHATE ION, ... | | Authors: | Aleshin, A.E, Liu, X, Kirby, C, Bourenkov, G.P, Bartunik, H.D, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1999-11-24 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of mutant monomeric hexokinase I reveal multiple ADP binding sites and conformational changes relevant to allosteric regulation.
J.Mol.Biol., 296, 2000
|
|
2EWF
 
 | | Crystal structure of the site-specific DNA nickase N.BspD6I | | Descriptor: | BROMIDE ION, Nicking endonuclease N.BspD6I | | Authors: | Kachalova, G.S, Bartunik, H.D, Artyukh, R.I, Rogulin, E.A, Perevyazova, T.A, Zheleznaya, L.A, Matvienko, N.I. | | Deposit date: | 2005-11-03 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of the heterodimeric type IIS restriction endonuclease R.BspD6I acting as a complex between a monomeric site-specific nickase and a catalytic subunit.
J.Mol.Biol., 384, 2008
|
|
2AU7
 
 | | The R43Q active site variant of E.coli inorganic pyrophosphatase | | Descriptor: | CHLORIDE ION, Inorganic pyrophosphatase, MANGANESE (II) ION, ... | | Authors: | Samygina, V.R, Avaeva, S.M, Bartunik, H.D. | | Deposit date: | 2005-08-27 | | Release date: | 2006-08-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Reversible inhibition of Escherichia coli inorganic pyrophosphatase by fluoride: trapped catalytic intermediates in cryo-crystallographic studies
J.Mol.Biol., 366, 2007
|
|
2AUU
 
 | | Inorganic pyrophosphatase complexed with magnesium pyrophosphate and fluoride | | Descriptor: | CHLORIDE ION, FLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Samygina, V.R, Popov, A.N, Avaeva, S.M, Bartunik, H.D. | | Deposit date: | 2005-08-29 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Reversible inhibition of Escherichia coli inorganic pyrophosphatase by fluoride: trapped catalytic intermediates in cryo-crystallographic studies
J.Mol.Biol., 366, 2007
|
|
2AU9
 
 | | Inorganic pyrophosphatase complexed with substrate | | Descriptor: | CHLORIDE ION, FLUORIDE ION, Inorganic pyrophosphatase, ... | | Authors: | Samygina, V.R, Popov, A.N, Avaeva, S.M, Bartunik, H.D. | | Deposit date: | 2005-08-27 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Reversible inhibition of Escherichia coli inorganic pyrophosphatase by fluoride: trapped catalytic intermediates in cryo-crystallographic studies
J.Mol.Biol., 366, 2007
|
|
2AU6
 
 | | Crystal structure of catalytic intermediate of inorganic pyrophosphatase | | Descriptor: | CHLORIDE ION, FLUORIDE ION, Inorganic pyrophosphatase, ... | | Authors: | Samygina, V.R, Popov, A.N, Avaeva, S.M, Bartunik, H.D. | | Deposit date: | 2005-08-27 | | Release date: | 2006-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Reversible inhibition of Escherichia coli inorganic pyrophosphatase by fluoride: trapped catalytic intermediates in cryo-crystallographic studies
J.Mol.Biol., 366, 2007
|
|
2AU8
 
 | | Catalytic intermediate structure of inorganic pyrophosphatase | | Descriptor: | CHLORIDE ION, Inorganic pyrophosphatase, MANGANESE (II) ION, ... | | Authors: | Samygina, V.R, Popov, A.N, Avaeva, S.M, Bartunik, H.D. | | Deposit date: | 2005-08-27 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Reversible inhibition of Escherichia coli inorganic pyrophosphatase by fluoride: trapped catalytic intermediates in cryo-crystallographic studies
J.Mol.Biol., 366, 2007
|
|
2BJB
 
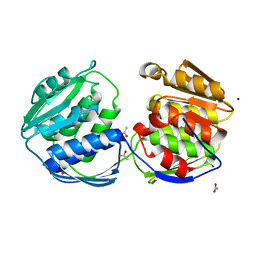 | | Mycobacterium Tuberculosis Epsp Synthase In Unliganded State | | Descriptor: | 3-PHOSPHOSHIKIMATE 1-CARBOXYVINYLTRANSFERASE, ACETATE ION, SODIUM ION | | Authors: | Bourenkov, G.P, Kachalova, G.S, Strizhov, N, Bruning, M, Vagin, A, Bartunik, H.D. | | Deposit date: | 2005-02-01 | | Release date: | 2006-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mycobacterium Tuberculosis Epsp Synthase in Unliganded State
To be Published
|
|
1HKB
 
 | | CRYSTAL STRUCTURE OF RECOMBINANT HUMAN BRAIN HEXOKINASE TYPE I COMPLEXED WITH GLUCOSE AND GLUCOSE-6-PHOSPHATE | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, CALCIUM ION, D-GLUCOSE 6-PHOSPHOTRANSFERASE, ... | | Authors: | Aleshin, A.E, Zeng, C, Burenkov, G.P, Bartunik, H.D, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1997-12-01 | | Release date: | 1998-06-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The mechanism of regulation of hexokinase: new insights from the crystal structure of recombinant human brain hexokinase complexed with glucose and glucose-6-phosphate.
Structure, 6, 1998
|
|
3PNY
 
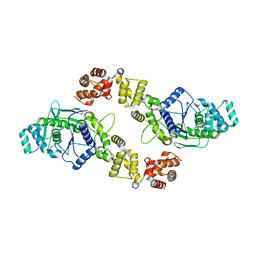 | |
3PNV
 
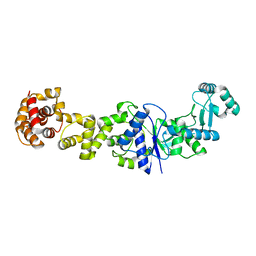 | |
1HH2
 
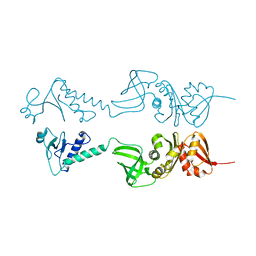 | | Crystal structure of NusA from Thermotoga maritima | | Descriptor: | N UTILIZATION SUBSTANCE PROTEIN A | | Authors: | Worbs, M, Bourenkov, G.P, Bartunik, H.D, Huber, R, Wahl, M.C. | | Deposit date: | 2000-12-18 | | Release date: | 2001-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Extended RNA Binding Surface Through Arrayed S1 and Kh Domains in Transcription Factor Nusa
Mol.Cell, 7, 2001
|
|
1JNR
 
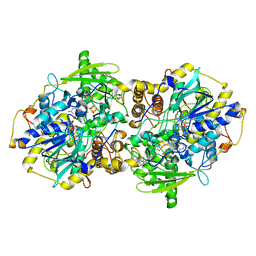 | | Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6 resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Fritz, G, Roth, A, Schiffer, A, Buechert, T, Bourenkov, G, Bartunik, H.D, Huber, H, Stetter, K.O, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2001-07-25 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JNZ
 
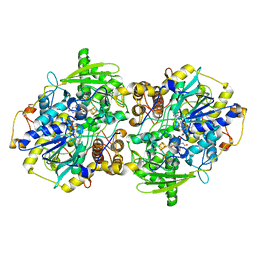 | | Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6 resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, SULFITE ION, ... | | Authors: | Fritz, G, Roth, A, Schiffer, A, Buechert, T, Bourenkov, G, Bartunik, H.D, Huber, H, Stetter, K.O, Kroneck, P.M, Ermler, U. | | Deposit date: | 2001-07-26 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LI1
 
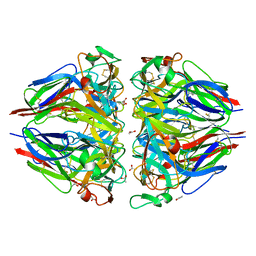 | | The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link | | Descriptor: | ACETATE ION, Collagen alpha 1(IV), Collagen alpha 2(IV) | | Authors: | Than, M.E, Henrich, S, Huber, R, Ries, A, Mann, K, Kuhn, K, Timpl, R, Bourenkov, G.P, Bartunik, H.D, Bode, W. | | Deposit date: | 2002-04-17 | | Release date: | 2002-05-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1RYP
 
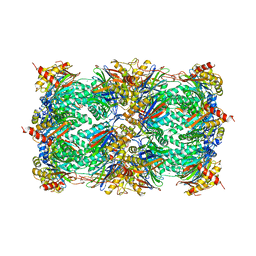 | | CRYSTAL STRUCTURE OF THE 20S PROTEASOME FROM YEAST AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | 20S PROTEASOME, MAGNESIUM ION | | Authors: | Groll, M, Ditzel, L, Loewe, J, Stock, D, Bochtler, M, Bartunik, H.D, Huber, R. | | Deposit date: | 1997-02-26 | | Release date: | 1998-04-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of 20S proteasome from yeast at 2.4 A resolution.
Nature, 386, 1997
|
|
1VET
 
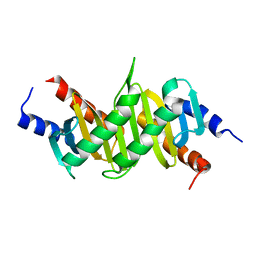 | | Crystal Structure of p14/MP1 at 1.9 A resolution | | Descriptor: | Late endosomal/lysosomal Mp1 interacting protein, Mitogen-activated protein kinase kinase 1 interacting protein 1 | | Authors: | Kurzbauer, R, Teis, D, Maurer-Stroh, S, Eisenhaber, F, Hekman, M, Bourenkov, G.P, Bartunik, H.D, Huber, L.A, Clausen, T. | | Deposit date: | 2004-04-05 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the p14/MP1 scaffolding complex: How a twin couple attaches mitogen- activated protein kinase signaling to late endosomes
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1VEU
 
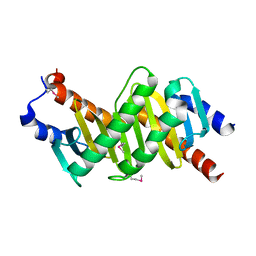 | | Crystal structure of the p14/MP1 complex at 2.15 A resolution | | Descriptor: | Late endosomal/lysosomal Mp1 interacting protein, Mitogen-activated protein kinase kinase 1 interacting protein 1 | | Authors: | Kurzbauer, R, Teis, D, Maurer-Stroh, S, Eisenhaber, F, Hekman, M, Bourenkov, G.P, Bartunik, H.D, Huber, L.A, Clausen, T. | | Deposit date: | 2004-04-05 | | Release date: | 2004-08-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the p14/MP1 scaffolding complex: How a twin couple attaches mitogen- activated protein kinase signaling to late endosomes
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1QV9
 
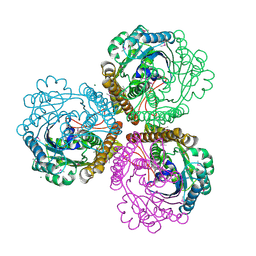 | | Coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase (Mtd) from Methanopyrus kandleri: A methanogenic enzyme with an unusual quarternary structure | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Hagemeier, C.H, Shima, S, Thauer, R.K, Bourenkov, G, Bartunik, H.D, Ermler, U. | | Deposit date: | 2003-08-27 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase (Mtd) from Methanopyrus kandleri: a methanogenic enzyme with an unusual quarternary structure
J.Mol.Biol., 332, 2003
|
|
1S0P
 
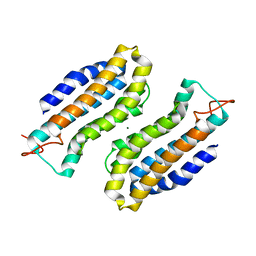 | | Structure of the N-Terminal Domain of the Adenylyl Cyclase-Associated Protein (CAP) from Dictyostelium discoideum. | | Descriptor: | Adenylyl cyclase-associated protein, MAGNESIUM ION | | Authors: | Ksiazek, D, Brandstetter, H, Israel, L, Bourenkov, G.P, Katchalova, G, Janssen, K.P, Bartunik, H.D, Noegel, A.A, Schleicher, M, Holak, T.A. | | Deposit date: | 2004-01-01 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | STRUCTURE OF THE N-TERMINAL DOMAIN OF THE ADENYLYL
CYCLASE-ASSOCIATED PROTEIN (CAP) FROM DICTYOSTELIUM DISCOIDEUM
Structure, 11, 2003
|
|
4DP1
 
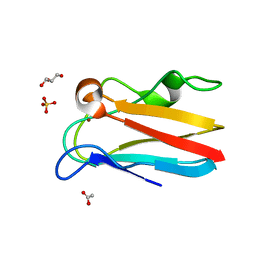 | | The 1.35 Angstrom crystal structure of reduced (CuI) poplar plastocyanin B at pH 4.0 | | Descriptor: | ACETATE ION, COPPER (I) ION, GLYCEROL, ... | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
4DP7
 
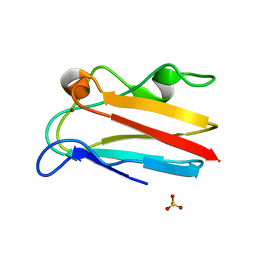 | | The 1.08 Angstrom crystal structure of oxidized (CuII) poplar plastocyanin A at pH 4.0 | | Descriptor: | COPPER (II) ION, Plastocyanin A, chloroplastic, ... | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
4DP4
 
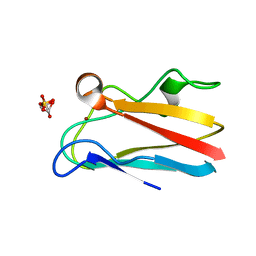 | | The 1.54 Angstrom crystal structure of reduced (CuI) poplar plastocyanin B at pH 6.0 | | Descriptor: | COPPER (I) ION, GLYCEROL, Plastocyanin B, ... | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
4DP6
 
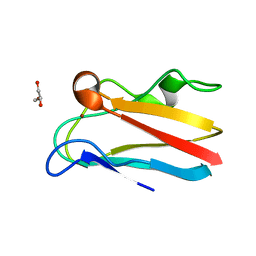 | | The 1.67 Angstrom crystal structure of reduced (CuI) poplar plastocyanin B at pH 8.0 | | Descriptor: | COPPER (I) ION, GLYCEROL, Plastocyanin B, ... | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
