2Q7O
 
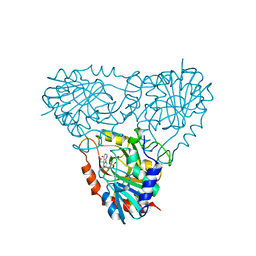 | | Structure of human purine nucleoside phosphorylase in complex with L-Immucillin-H | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Rinaldo-Matthis, A, Almo, S.C, Schramm, V.L. | | Deposit date: | 2007-06-07 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | L-Enantiomers of transition state analogue inhibitors bound to human purine nucleoside phosphorylase.
J.Am.Chem.Soc., 130, 2008
|
|
4WUT
 
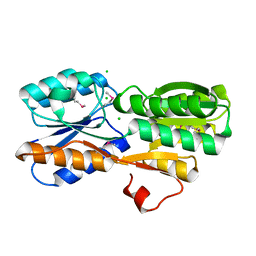 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE | | Descriptor: | ABC transporter substrate binding protein (Ribose), CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-03 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS (Avi_5133, TARGET EFI-511220) WITH BOUND D-FUCOSE
To be published
|
|
2QYA
 
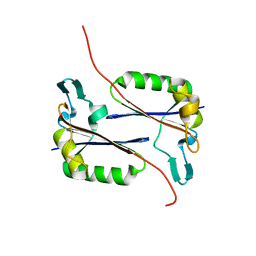 | | Crystal structure of an uncharacterized conserved protein from Methanopyrus kandleri | | Descriptor: | Uncharacterized conserved protein | | Authors: | Bonanno, J.B, Zhang, A, Bain, K.T, Adams, J, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-14 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of an uncharacterized conserved protein from Methanopyrus kandleri.
To be Published
|
|
2QDN
 
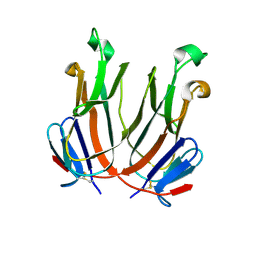 | |
2QT3
 
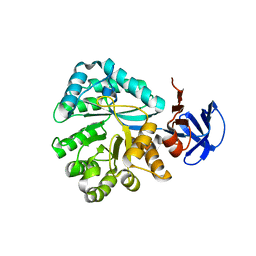 | | Crystal structure of N-Isopropylammelide isopropylaminohydrolase AtzC from Pseudomonas sp. strain ADP complexed with Zn | | Descriptor: | N-isopropylammelide isopropyl amidohydrolase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Seffernick, J, Wackett, L.P, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-01 | | Release date: | 2007-09-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structure of N-Isopropylammelide isopropylaminohydrolase AtzC from Pseudomonas sp. strain ADP complexed with Zn.
To be Published
|
|
2QY6
 
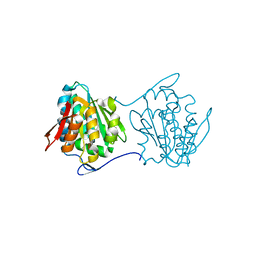 | | Crystal structure of the N-terminal domain of UPF0209 protein yfcK from Escherichia coli O157:H7 | | Descriptor: | UPF0209 protein yfcK | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Eberle, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-13 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the N-terminal domain of UPF0209 protein yfcK from Escherichia coli O157:H7.
To be Published
|
|
4WT7
 
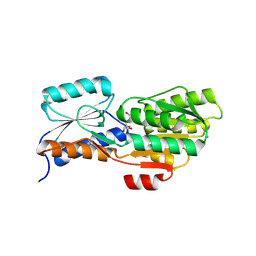 | | Crystal structure of an ABC transporter solute binding protein (IPR025997) from Agrobacterium vitis (Avi_5165, Target EFI-511223) with bound allitol | | Descriptor: | ABC transporter substrate binding protein (Ribose), CHLORIDE ION, D-allitol | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-29 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an ABC transporter solute binding protein (IPR025997) from Agrobacterium vitis (Avi_5165, Target EFI-511223) with bound allitol
To be published
|
|
2QF9
 
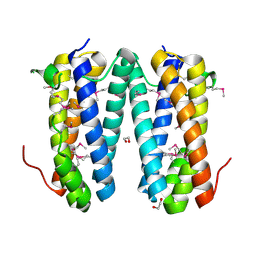 | | Crystal structure of putative secreted protein DUF305 from Streptomyces coelicolor | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein | | Authors: | Ramagopal, U.A, Rutter, M, Adams, J, Toro, R, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-06-27 | | Release date: | 2007-07-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure of putative secreted protein DUF305 from Streptomyces coelicolor.
To be Published
|
|
2QE3
 
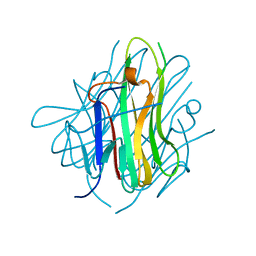 | | Crystal structure of human tl1a extracellular domain | | Descriptor: | CHLORIDE ION, TNF superfamily ligand TL1A | | Authors: | Zhan, C, Yan, Q, Patskovsky, Y, Shi, W, Toro, R, Bonanno, J, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-06-22 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical and structural characterization of the human TL1A ectodomain.
Biochemistry, 48, 2009
|
|
2QEE
 
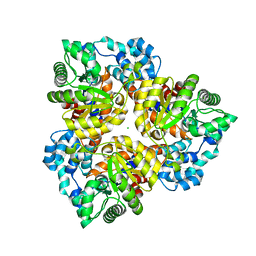 | | Crystal structure of putative amidohydrolase BH0493 from Bacillus halodurans C-125 | | Descriptor: | BH0493 protein, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-06-25 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of putative amidohydrolase BH0493 from Bacillus halodurans C-125.
To be Published
|
|
2QJJ
 
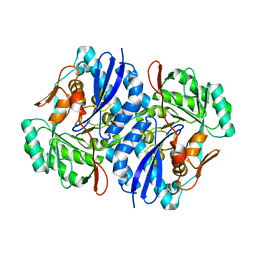 | | Crystal structure of D-Mannonate dehydratase from Novosphingobium aromaticivorans | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Vick, J.E, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-07-07 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: D-Mannonate dehydratase from Novosphingobium aromaticivorans.
Biochemistry, 46, 2007
|
|
2QKP
 
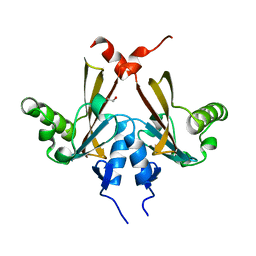 | | Crystal structure of C-terminal domain of SMU_1151c from Streptococcus mutans | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Uncharacterized protein | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Wu, B, Bain, K, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-11 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of C-terminal domain of SMU_1151c from Streptococcus mutans.
To be Published
|
|
2QDD
 
 | | Crystal structure of a member of enolase superfamily from Roseovarius nubinhibens ISM | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Patskovsky, Y, Bonanno, J, Sauder, J.M, Gilmore, J.M, Iizuka, M, Groshong, C, Gheyi, T, Sojitra, S, Wasserman, S.R, Koss, J, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-06-20 | | Release date: | 2007-06-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a member of enolase superfamily from Roseovarius nubinhibens ISM.
To be Published
|
|
4WZZ
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM CLOSTRIDIUM PHYTOFERMENTAS (Cphy_0583, TARGET EFI-511148) WITH BOUND L-RHAMNOSE | | Descriptor: | 1,2-ETHANEDIOL, Putative sugar ABC transporter, substrate-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-20 | | Release date: | 2014-12-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM CLOSTRIDIUM PHYTOFERMENTAS (Cphy_0583, TARGET EFI-511148) WITH BOUND L-RHAMNOSE
To be published
|
|
2QR4
 
 | | Crystal structure of oligoendopeptidase-F from Enterococcus faecium | | Descriptor: | Peptidase M3B, oligoendopeptidase F | | Authors: | Ramagopal, U.A, Toro, R, Meyer, A.J, Freeman, J, Bain, K, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-14 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of oligoendopeptidase-F from Enterococcus faecium.
To be Published
|
|
2QYZ
 
 | | Crystal structure of the uncharacterized protein CTC02137 from Clostridium tetani E88 | | Descriptor: | Uncharacterized protein | | Authors: | Malashkevich, V.N, Toro, R, Meyer, A.J, Sauder, J.M, Wasserman, T, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-15 | | Release date: | 2007-08-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of the uncharacterized protein CTC02137 from Clostridium tetani E88.
To be Published
|
|
4WR2
 
 | | Crystal structure of a putative pyrimidine-specific ribonucleoside hydrolase (RihA) Protein from Shewanella loihica PV-4 (SHEW_0697, Target PSI-029635) with divalent cation and PEG 400 bound at the active site | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, Pyrimidine-specific ribonucleoside hydrolase RihA | | Authors: | Himmel, D.M, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R.D, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a putative pyrimidine-specific ribonucleoside hydrolase (RihA) Protein from Shewanella loihica PV-4 (SHEW_0697, Target PSI-029635) with divalent cation and PEG 400 bound at the active site
To be published
|
|
4WWH
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEG_1704, TARGET EFI-510967) WITH BOUND D-GALACTOSE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ABC transporter, beta-D-galactopyranose | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEG_1704, TARGET EFI-510967) WITH BOUND D-GALACTOSE
To be published
|
|
2QWT
 
 | | Crystal structure of the TetR transcription regulatory protein from Mycobacterium vanbaalenii | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Adams, J, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-10 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the TetR transcription regulatory protein from Mycobacterium vanbaalenii.
To be Published
|
|
4X8R
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM Rhodobacter sphaeroides (Rsph17029_2138, TARGET EFI-510205) WITH BOUND Glucuronate | | Descriptor: | PHOSPHATE ION, TRAP dicarboxylate transporter, DctP subunit, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-10 | | Release date: | 2014-12-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM Rhodobacter sphaeroides (Rsph17029_2138, TARGET EFI-510205) WITH BOUND Glucuronate
To be published
|
|
2QJN
 
 | | Crystal structure of D-mannonate dehydratase from Novosphingobium aromaticivorans complexed with Mg and 2-keto-3-deoxy-D-gluconate | | Descriptor: | 2-KETO-3-DEOXYGLUCONATE, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Vick, J.E, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-07-08 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: D-Mannonate dehydratase from Novosphingobium aromaticivorans.
Biochemistry, 46, 2007
|
|
2QGY
 
 | | Crystal structure of an enolase from the environmental genome shotgun sequencing of the Sargasso Sea | | Descriptor: | Enolase from the environmental genome shotgun sequencing of the Sargasso Sea, MAGNESIUM ION | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Lau, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an enolase from the environmental genome shotgun sequencing of the Sargasso Sea.
To be Published
|
|
4WUV
 
 | | Crystal Structure of a putative D-Mannonate oxidoreductase from Haemophilus influenza (Avi_5165, TARGET EFI-513796) with bound NAD | | Descriptor: | 1,2-ETHANEDIOL, 2-hydroxycyclohexanecarboxyl-CoA dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2014-11-03 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Crystal structure of a putative D-Mannonate oxidoreductase from Haemophilus influenza (Avi_5165, TARGET EFI-513796) with bound NAD
To be published
|
|
4WXE
 
 | | CRYSTAL STRUCTURE OF A LACI REGULATOR FROM LACTOBACILLUS CASEI (LSEI_2103, TARGET EFI-512911) WITH BOUND TRIS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-13 | | Release date: | 2014-11-26 | | Last modified: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF A LACI REGULATOR FROM LACTOBACILLUS CASEI (LSEI_2103, TARGET EFI-512911) WITH BOUND TRIS
To be published
|
|
2QU7
 
 | | Crystal structure of a putative transcription regulator from Staphylococcus saprophyticus subsp. saprophyticus | | Descriptor: | CHLORIDE ION, Putative transcriptional regulator | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Mendoza, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-03 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative transcription regulator from Staphylococcus saprophyticus subsp. saprophyticus.
To be Published
|
|
