2CV8
 
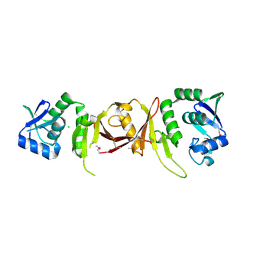 | |
2CSO
 
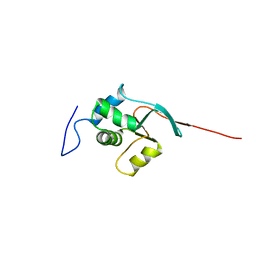 | |
2CU1
 
 | |
2CZ9
 
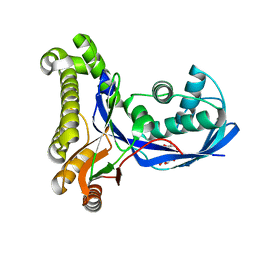 | | Crystal Structure of galactokinase from Pyrococcus horikoshi | | Descriptor: | CHLORIDE ION, GLYCEROL, Probable galactokinase | | Authors: | Inagaki, E, Sakamoto, K, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-12 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of galactokinase from Pyrococcus horikoshi
To be Published
|
|
2CXA
 
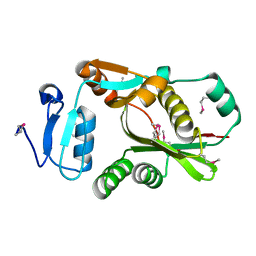 | | Crystal structure of Leucyl/phenylalanyl-tRNA protein transferase from Escherichia coli | | Descriptor: | Leucyl/phenylalanyl-tRNA-protein transferase | | Authors: | Kato-Murayama, M, Bessho, Y, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-28 | | Release date: | 2005-12-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of leucyl/phenylalanyl-tRNA-protein transferase from Escherichia coli
Protein Sci., 16, 2007
|
|
2CYY
 
 | | Crystal structure of PH1519 from Pyrococcus horikosii OT3 | | Descriptor: | CALCIUM ION, GLUTAMINE, Putative HTH-type transcriptional regulator PH1519 | | Authors: | Okazaki, N, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-08 | | Release date: | 2006-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of PH1519 from Pyrococcus horikosii OT3
To be Published
|
|
6ACR
 
 | | Crystal structure of human ALK2 kinase domain with R206H mutation in complex with RK-59638 | | Descriptor: | Activin receptor type-1, N-(4-methoxyphenyl)-4-[3-(pyridin-3-yl)-1H-pyrazol-4-yl]pyrimidin-2-amine, SULFATE ION | | Authors: | Sakai, N, Mishima-Tsumagari, C, Matsumoto, T, Shirouzu, M. | | Deposit date: | 2018-07-27 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Bis-Heteroaryl Pyrazoles: Identification of Orally Bioavailable Inhibitors of Activin Receptor-Like Kinase-2 (R206H).
Chem. Pharm. Bull., 67, 2019
|
|
4Z16
 
 | | Crystal Structure of the Jak3 Kinase Domain Covalently Bound to N-(3-(((5-chloro-2-((2-methoxy-4-(4-methylpiperazin-1-yl)phenyl)amino)pyrimidin-4-yl)amino)methyl)phenyl)acrylamide | | Descriptor: | N-(3-{[(5-chloro-2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]methyl}phenyl)prop-2-enamide, Tyrosine-protein kinase JAK3 | | Authors: | McNally, R, Tan, L, Gray, N.S, Eck, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of Selective Covalent Janus Kinase 3 Inhibitors.
J.Med.Chem., 58, 2015
|
|
6JUX
 
 | | Crystal structure of human ALK2 kinase domain with R206H mutation in complex with RK-71807 | | Descriptor: | 4-(1-ethyl-3-pyridin-3-yl-pyrazol-4-yl)-~{N}-(4-piperazin-1-ylphenyl)pyrimidin-2-amine, Activin receptor type-1, SULFATE ION | | Authors: | Sakai, N, Mishima-Tsumagari, C, Matsumoto, T, Shirouzu, M. | | Deposit date: | 2019-04-15 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Activin Receptor-Like Kinase 2 (R206H) Inhibition by Bis-heteroaryl Pyrazole-Based Inhibitors for the Treatment of Fibrodysplasia Ossificans Progressiva Identified by the Integration of Ligand-Based and Structure-Based Drug Design Approaches.
Acs Omega, 5, 2020
|
|
1RKR
 
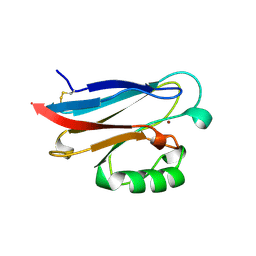 | | CRYSTAL STRUCTURE OF AZURIN-I FROM ALCALIGENES XYLOSOXIDANS NCIMB 11015 | | Descriptor: | AZURIN-I, COPPER (II) ION | | Authors: | Li, C, Inoue, T, Gotowda, M, Suzuki, S, Yamaguchi, K, Kataoka, K, Kai, Y. | | Deposit date: | 1997-05-17 | | Release date: | 1998-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of azurin I from the denitrifying bacterium Alcaligenes xylosoxidans NCIMB 11015 at 2.45 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
5VUB
 
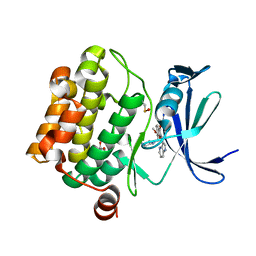 | | Pim1 Kinase in complex with a benzofuranone inhibitor | | Descriptor: | (2Z)-6-methoxy-7-(piperazin-1-ylmethyl)-2-(1H-pyrrolo[3,2-b]pyridin-3-ylmethylidene)-1-benzofuran-3-one, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Parker, L.J. | | Deposit date: | 2017-05-18 | | Release date: | 2017-12-13 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Theoretical Analysis of Activity Cliffs among Benzofuranone-Class Pim1 Inhibitors Using the Fragment Molecular Orbital Method with Molecular Mechanics Poisson-Boltzmann Surface Area (FMO+MM-PBSA) Approach
J Chem Inf Model, 57, 2017
|
|
2GS9
 
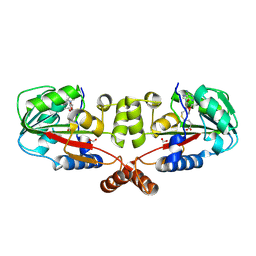 | | Crystal structure of TT1324 from Thermus thermophilis HB8 | | Descriptor: | FORMIC ACID, Hypothetical protein TT1324, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kamitori, S, Abe, A, Ebihara, A, Kanagawa, M, Nakagawa, N, Kuroishi, C, Agari, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-25 | | Release date: | 2007-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of TT1324 from Thermus thermophilis HB8
To be Published
|
|
1E7Z
 
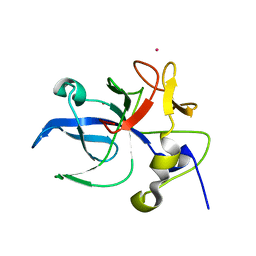 | | Crystal structure of the EMAP2/RNA binding domain of the p43 protein from human aminoacyl-tRNA synthetase complex | | Descriptor: | ENDOTHELIAL-MONOCYTE ACTIVATING POLYPEPTIDE II, MERCURY (II) ION | | Authors: | Pasqualato, S, Kerjan, P, Renault, L, Menetrey, J, Mirande, M, Cherfils, J. | | Deposit date: | 2000-09-13 | | Release date: | 2000-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the Emapii Domain of Human Aminoacyl-tRNA Synthetase Complex Reveals Evolutionary Dimeric Mimicry
Embo J., 20, 2001
|
|
7DEV
 
 | | Crystal Structures of Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora | | Descriptor: | Anthocyanin 5-aromatic acyltransferase | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2020-11-05 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora, a structural insight into biosynthesis of a blue anthocyanin.
Phytochemistry, 186, 2021
|
|
7DEX
 
 | | Crystal Structures of Anthocyanin 5,3'-aromatic acyltransferase H174A mutant with caffeoyl-CoA | | Descriptor: | Anthocyanin 5-aromatic acyltransferase, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (E)-3-[3,4-bis(oxidanyl)phenyl]prop-2-enethioate | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2020-11-05 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora, a structural insight into biosynthesis of a blue anthocyanin.
Phytochemistry, 186, 2021
|
|
5K7X
 
 | |
5XON
 
 | | RNA Polymerase II elongation complex bound with Spt4/5 and TFIIS | | Descriptor: | DNA (48-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ehara, H, Yokoyama, T, Shigematsu, H, Shirouzu, M, Sekine, S. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structure of the complete elongation complex of RNA polymerase II with basal factors
Science, 357, 2017
|
|
2YVQ
 
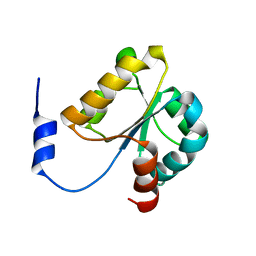 | | Crystal structure of MGS domain of carbamoyl-phosphate synthetase from homo sapiens | | Descriptor: | Carbamoyl-phosphate synthase | | Authors: | Xie, Y, Ihsanawati, Kishishita, S, Murayama, K, Takemoto, C, Shirozu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-13 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of MGS domain of carbamoyl-phosphate synthetase from homo sapiens
To be Published
|
|
3P0B
 
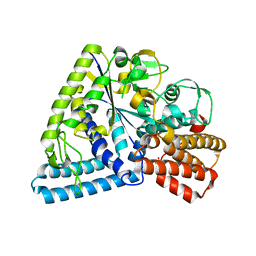 | |
4E1B
 
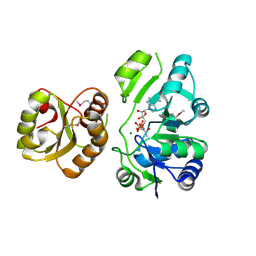 | | Re-refinement of PDB entry 2EQA - SUA5 protein from Sulfolobus tokodaii with bound threonylcarbamoyladenylate | | Descriptor: | MAGNESIUM ION, YrdC/Sua5 family protein, threonylcarbamoyladenylate | | Authors: | Parthier, C, Goerlich, S, Jaenecke, F, Breithaupt, C, Braeuer, U, Fandrich, U, Clausnitzer, D, Wehmeier, U.F, Boettcher, C, Scheel, D, Stubbs, M.T. | | Deposit date: | 2012-03-06 | | Release date: | 2012-03-14 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The O-Carbamoyltransferase TobZ Catalyzes an Ancient Enzymatic Reaction.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
5VUA
 
 | | Pim1 Kinase in complex with a benzofuranone inhibitor | | Descriptor: | (2Z)-6-methoxy-7-(piperazin-1-ylmethyl)-2-(1H-pyrrolo[2,3-c]pyridin-3-ylmethylidene)-1-benzofuran-3-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Parker, L.J. | | Deposit date: | 2017-05-18 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Theoretical Analysis of Activity Cliffs among Benzofuranone-Class Pim1 Inhibitors Using the Fragment Molecular Orbital Method with Molecular Mechanics Poisson-Boltzmann Surface Area (FMO+MM-PBSA) Approach
J Chem Inf Model, 57, 2017
|
|
5VUC
 
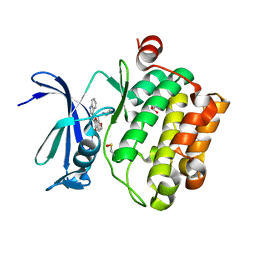 | | Pim1 Kinase in complex with a benzofuranone inhibitor | | Descriptor: | (2Z)-2-(1H-indol-3-ylmethylidene)-6-methoxy-7-(piperazin-1-ylmethyl)-1-benzofuran-3-one, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Parker, L.J. | | Deposit date: | 2017-05-18 | | Release date: | 2017-12-13 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Theoretical Analysis of Activity Cliffs among Benzofuranone-Class Pim1 Inhibitors Using the Fragment Molecular Orbital Method with Molecular Mechanics Poisson-Boltzmann Surface Area (FMO+MM-PBSA) Approach
J Chem Inf Model, 57, 2017
|
|
2YRW
 
 | | Crystal structure of GAR synthetase from Geobacillus kaustophilus | | Descriptor: | PHOSPHATE ION, Phosphoribosylglycinamide synthetase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
2YS6
 
 | | Crystal structure of GAR synthetase from Geobacillus kaustophilus | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCINE, Phosphoribosylglycinamide synthetase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
2YS7
 
 | | Crystal structure of GAR synthetase from Geobacillus kaustophilus | | Descriptor: | Phosphoribosylglycinamide synthetase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
