4MX1
 
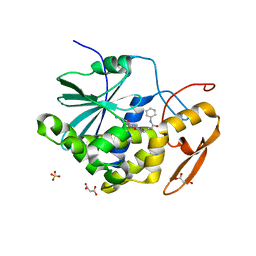 | | Structure of ricin A chain bound with 2-amino-4-oxo-N-(2-(3-phenylureido)ethyl)-3,4-dihydropteridine-7-carboxamide | | Descriptor: | 2-amino-4-oxo-N-{2-[(phenylcarbamoyl)amino]ethyl}-3,4-dihydropteridine-7-carboxamide, MALONIC ACID, Ricin A chain, ... | | Authors: | Robertus, J.D, Wiget, P.A, Manzano, L.A, Pruet, J.M, Gao, G, Saito, R, Jasheway, K.R, Monzingo, A.F, Anslyn, E.V. | | Deposit date: | 2013-09-25 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Sulfur incorporation generally improves Ricin inhibition in pterin-appended glycine-phenylalanine dipeptide mimics.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2Y5S
 
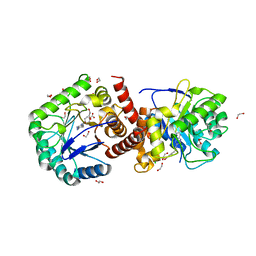 | | Crystal structure of Burkholderia cenocepacia dihydropteroate synthase complexed with 7,8-dihydropteroate. | | Descriptor: | 1,2-ETHANEDIOL, 7,8-DIHYDROPTEROATE, CHLORIDE ION, ... | | Authors: | Morgan, R.E, Batot, G.O, Dement, J.M, Rao, V.A, Eadsforth, T.C, Hunter, W.N. | | Deposit date: | 2011-01-17 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Burkholderia Cenocepacia Dihydropteroate Synthase in the Apo-Form and Complexed with the Product 7,8-Dihydropteroate.
Bmc Struct.Biol., 11, 2011
|
|
4N4A
 
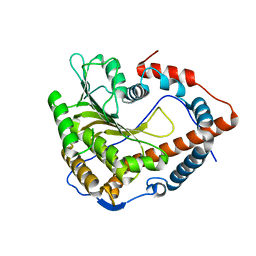 | | Cystal structure of Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 | | Authors: | Smietanski, M, Werener, M, Purta, E, Kaminska, K.H, Stepinski, J, Darzynkiewicz, E, Nowotny, M, Bujnicki, J.M. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural analysis of human 2'-O-ribose methyltransferases involved in mRNA cap structure formation.
Nat Commun, 5, 2014
|
|
4OQB
 
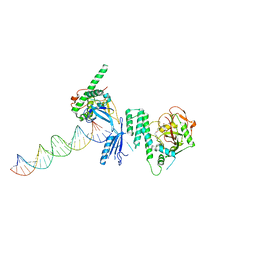 | | Structure of Human PARP-1 bound to a DNA double strand break in complex with (2Z)-2-{4-[2-(morpholin-4-yl)ethoxy]benzylidene}-3-oxo-2,3-dihydro-1-benzofuran-7-carboxamide | | Descriptor: | (2Z)-2-{4-[2-(morpholin-4-yl)ethoxy]benzylidene}-3-oxo-2,3-dihydro-1-benzofuran-7-carboxamide, DNA (26-MER), Poly [ADP-ribose] polymerase 1, ... | | Authors: | Pascal, J.M, Steffen, J.D. | | Deposit date: | 2014-02-07 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.362 Å) | | Cite: | Discovery and Structure-Activity Relationship of Novel 2,3-Dihydrobenzofuran-7-carboxamide and 2,3-Dihydrobenzofuran-3(2H)-one-7-carboxamide Derivatives as Poly(ADP-ribose)polymerase-1 Inhibitors.
J.Med.Chem., 57, 2014
|
|
4ON0
 
 | | Crystal Structure of NolR from Sinorhizobium fredii in complex with oligo AA DNA | | Descriptor: | DNA (5 -D(*TP*AP*AP*TP*CP*TP*CP*TP*TP*GP*GP*GP*AP*CP*TP*AP*CP*AP*AP*TP*TP*A)-3 ), DNA (5 -D(*TP*AP*TP*TP*AP*GP*AP*GP*AP*AP*CP*CP*CP*TP*GP*AP*TP*GP*TP*TP*AP*A)-3 ), NolR | | Authors: | Lee, S.G, Krishnan, H.B, Jez, J.M. | | Deposit date: | 2014-01-28 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for regulation of rhizobial nodulation and symbiosis gene expression by the regulatory protein NolR.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2YJ8
 
 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | Descriptor: | (2S,4R)-4-(2-chlorophenyl)sulfonyl-N-[1-(iminomethyl)cyclopropyl]-1-[1-(4-iodophenyl)cyclopropyl]carbonyl-pyrrolidine-2-carboxamide, CATHEPSIN L1, GLYCEROL | | Authors: | Banner, D.W, Benz, J.M, Haap, W. | | Deposit date: | 2011-05-19 | | Release date: | 2011-11-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Halogen Bonding at the Active Sites of Human Cathepsin L and Mek1 Kinase: Efficient Interactions in Different Environments.
Chemmedchem, 6, 2011
|
|
2Y7C
 
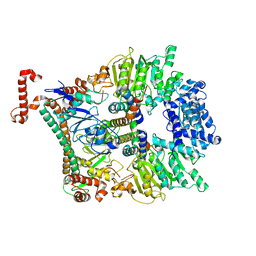 | | Atomic model of the Ocr-bound methylase complex from the Type I restriction-modification enzyme EcoKI (M2S1). Based on fitting into EM map 1534. | | Descriptor: | GENE 0.3 PROTEIN, TYPE I RESTRICTION ENZYME ECOKI M PROTEIN, TYPE-1 RESTRICTION ENZYME ECOKI SPECIFICITY PROTEIN | | Authors: | Kennaway, C.K, Obarska-Kosinska, A, White, J.H, Tuszynska, I, Cooper, L.P, Bujnicki, J.M, Trinick, J, Dryden, D.T.F. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | The Structure of M.Ecoki Type I DNA Methyltransferase with a DNA Mimic Antirestriction Protein.
Nucleic Acids Res., 37, 2009
|
|
2YBD
 
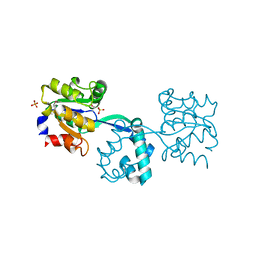 | | Crystal structure of probable had family hydrolase from pseudomonas fluorescens pf-5 with bound phosphate | | Descriptor: | HYDROLASE, HALOACID DEHALOGENASE-LIKE FAMILY, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Probable Had Family Hydrolase from Pseudomonas Fluorescens Pf-5 with Bound Phosphate
To be Published
|
|
2YIH
 
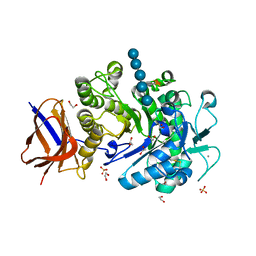 | | Structure of a Paenibacillus polymyxa Xyloglucanase from GH family 44 with Xyloglucan | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CEL44C, ... | | Authors: | Ariza, A, Eklof, J.M, Spadiut, O, Offen, W.A, Roberts, S.M, Besenmatter, W, Friis, E.P, Skjot, M, Wilson, K.S, Brumer, H, Davies, G. | | Deposit date: | 2011-05-13 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Activity of Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44.
J.Biol.Chem., 286, 2011
|
|
4P2B
 
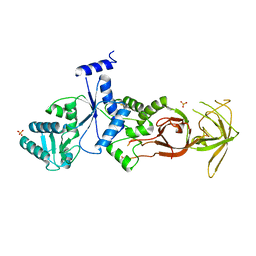 | |
2YJB
 
 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | Descriptor: | (2S,4R)-4-(2-chlorophenyl)sulfonyl-N-[1-(iminomethyl)cyclopropyl]-1-[1-[4-(trifluoromethyl)phenyl]cyclopropyl]carbonyl-pyrrolidine-2-carboxamide, CATHEPSIN L1, GLYCEROL | | Authors: | Banner, D.W, Benz, J.M, Haap, W. | | Deposit date: | 2011-05-19 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Halogen Bonding at the Active Sites of Human Cathepsin L and Mek1 Kinase: Efficient Interactions in Different Environments.
Chemmedchem, 6, 2011
|
|
4NMN
 
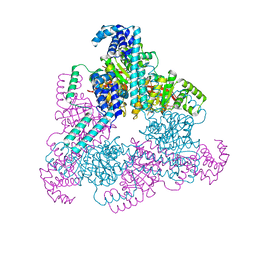 | | Aquifex aeolicus replicative helicase (DnaB) complexed with ADP, at 3.3 resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Replicative DNA helicase, ... | | Authors: | Lyubimov, A.Y, Strycharska, M.S, Erzberger, J.P, Berger, J.M. | | Deposit date: | 2013-11-15 | | Release date: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Nucleotide and partner-protein control of bacterial replicative helicase structure and function.
Mol.Cell, 52, 2013
|
|
4NQ5
 
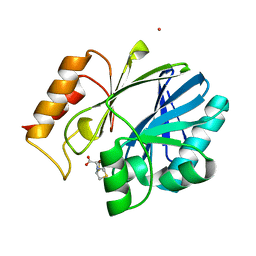 | | Bacillus cereus Zn-dependent metallo-beta-lactamase at pH 7 complexed with compound CS319 | | Descriptor: | (3R,5R,7aS)-5-(sulfanylmethyl)tetrahydro[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, Beta-lactamase 2, POTASSIUM ION, ... | | Authors: | Gonzalez, J.M, Gonzalez, M.M, Vila, A.J. | | Deposit date: | 2013-11-23 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Inhibition of metallo-lactamases with bicyclic compounds
TO BE PUBLISHED
|
|
4NX4
 
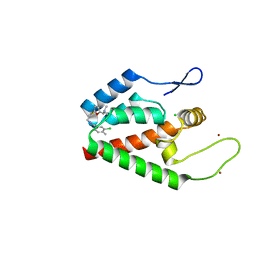 | | Re-refinement of CAP-1 HIV-CA complex | | Descriptor: | 1-(3-chloro-4-methylphenyl)-3-{2-[({5-[(dimethylamino)methyl]-2-furyl}methyl)thio]ethyl}urea, CHLORIDE ION, Gag-Pol polyprotein, ... | | Authors: | Lang, P.T, Holton, J.M, Fraser, J.S, Alber, T. | | Deposit date: | 2013-12-08 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protein structural ensembles are revealed by redefining X-ray electron density noise.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NDN
 
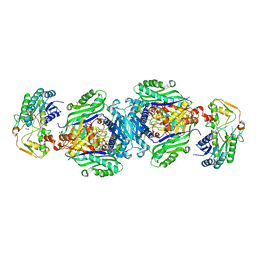 | | Structural insights of MAT enzymes: MATa2b complexed with SAM and PPNP | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Murray, B, Antonyuk, S.V, Marina, A, Lu, S.C, Mato, J.M, Hasnain, S.S, Rojas, A.L. | | Deposit date: | 2013-10-27 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and function study of the complex that synthesizes S-adenosylmethionine.
IUCrJ, 1, 2014
|
|
4NDB
 
 | | X-ray structure of a mutant (T61D) of calexcitin - a neuronal calcium-signalling protein | | Descriptor: | CALCIUM ION, Calexcitin | | Authors: | Erskine, P.T, Fokas, A, Muriithi, C, Razzall, E, Bowyer, A, Findlow, I.S, Werner, J.M, Wallace, B.A, Wood, S.P, Cooper, J.B. | | Deposit date: | 2013-10-25 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray, spectroscopic and normal-mode dynamics of calexcitin: structure-function studies of a neuronal calcium-signalling protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4P37
 
 | | Crystal structure of the Megavirus polyadenylate synthase | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Priet, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2014-03-06 | | Release date: | 2015-04-01 | | Last modified: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | mRNA maturation in giant viruses: variation on a theme.
Nucleic Acids Res., 43, 2015
|
|
4LRW
 
 | | Crystal Structure of K-Ras G12C (cysteine-light), GDP-bound | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-07-21 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4LQR
 
 | | Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 31/fibronectin type III domain protein | | Authors: | Grondin, J.M, Furness, H.S, Duan, D, Spencer, C.A, Allingham, J.S, Smith, S.P. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
Plos One, 12, 2017
|
|
4LXK
 
 | | Crystal Structure of Human Beta Secretase in Complex with compound 11d | | Descriptor: | (1R,3S,4S,5R)-3-(4-amino-3-fluoro-5-{[(2R)-1,1,1-trifluoro-3-methoxypropan-2-yl]oxy}benzyl)-5-[(3-tert-butylbenzyl)amino]tetrahydro-2H-thiopyran-4-ol 1-oxide, Beta-secretase 1, GLYCEROL | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of cyclic sulfoxide hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4M1S
 
 | | Crystal Structure of small molecule vinylsulfonamide 13 covalently bound to K-Ras G12C | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, K-Ras GTPase, N-{1-[N-(2,4-dichlorophenyl)glycyl]piperidin-4-yl}ethanesulfonamide | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-08-04 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4M29
 
 | | Structure of a GH39 Beta-xylosidase from Caulobacter crescentus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-xylosidase | | Authors: | Polo, C.C, Santos, C.R, Correa, J.M, Simao, R.C.G, Seixas, F.A.V, Murakami, M.T. | | Deposit date: | 2013-08-05 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a GH39 Beta-xylosidase from Caulobacter crescentus
Thesis
|
|
4M7I
 
 | | Crystal Structure of GSK6157 Bound to PERK (R587-R1092, delete A660-T867) at 2.34A Resolution | | Descriptor: | 1-[5-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-4-fluoro-1H-indol-1-yl]-2-(6-methylpyridin-2-yl)ethanone, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Gampe, R.T, Axten, J.M. | | Deposit date: | 2013-08-12 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery of 5-{4-fluoro-1-[(6-methyl-2-pyridinyl)acetyl]-2,3-dihydro-1H-indol-5-yl}-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2656157), a Potent and Selective PERK Inhibitor Selected for Preclinical Development
To be Published
|
|
4LZD
 
 | | Human DNA polymerase mu- Apoenzyme | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-directed DNA/RNA polymerase mu, ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2013-07-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Sustained active site rigidity during synthesis by human DNA polymerase mu.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4P5Y
 
 | | Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with N-acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, Glycosyl hydrolase, ... | | Authors: | Grondin, J.M, Allingham, J.S, Boraston, A.B, Smith, S.P. | | Deposit date: | 2014-03-04 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
PLoS ONE, 12, 2017
|
|
