4EIW
 
 | | Whole cytosolic region of atp-dependent metalloprotease FtsH (G399L) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH | | Authors: | Suno, R, Niwa, H, Tsuchiya, D, Yoshida, M, Morikawa, K. | | Deposit date: | 2012-04-06 | | Release date: | 2012-06-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of the whole cytosolic region of ATP-dependent protease FtsH
Mol.Cell, 22, 2006
|
|
1Q5X
 
 | | Structure of OF RRAA (MENG), a protein inhibitor of RNA processing | | Descriptor: | REGULATOR OF RNASE E ACTIVITY A | | Authors: | Monzingo, A.F, Gao, J, Qiu, J, Georgiou, G, Robertus, J.D. | | Deposit date: | 2003-08-11 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray Structure of Escherichia coli RraA (MenG), A Protein Inhibitor of RNA Processing.
J.Mol.Biol., 332, 2003
|
|
6IVA
 
 | |
5LBV
 
 | | Structural basis of zika and dengue virus potent antibody cross-neutralization | | Descriptor: | SODIUM ION, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, envelope protein E | | Authors: | Barba-Spaeth, G. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of potent Zika-dengue virus antibody cross-neutralization.
Nature, 536, 2016
|
|
6IIJ
 
 | | Cryo-EM structure of CV-A10 mature virion | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Chen, J.H, Ye, X.H, Cong, Y, Huang, Z. | | Deposit date: | 2018-10-06 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Coxsackievirus A10 atomic structure facilitating the discovery of a broad-spectrum inhibitor against human enteroviruses.
Cell Discov, 5, 2019
|
|
7MOA
 
 | | Cryo-EM structure of the c-MET II/HGF I complex bound with HGF II in a rigid conformation | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MOB
 
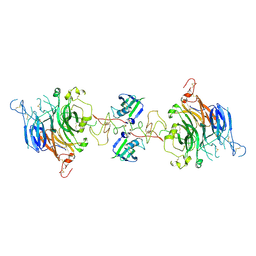 | | Cryo-EM structure of 2:2 c-MET/NK1 complex | | Descriptor: | Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MO8
 
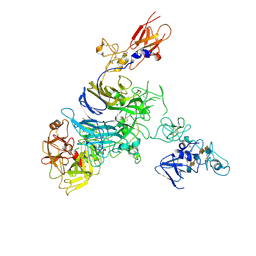 | | Cryo-EM structure of 1:1 c-MET I/HGF I complex after focused 3D refinement of holo-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MO9
 
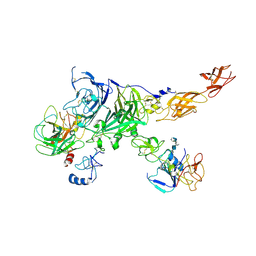 | | Cryo-EM map of the c-MET II/HGF I/HGF II (K4 and SPH) sub-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MO7
 
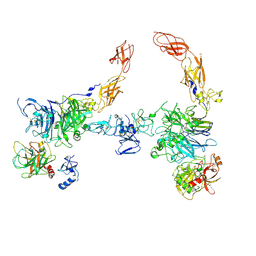 | | Cryo-EM structure of 2:2 c-MET/HGF holo-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
4H1M
 
 | | Crystal structure of PYK2 with the indole 10c | | Descriptor: | 7-({[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]carbamoyl}amino)-N-(propan-2-yl)-1H-indole-2-carboxamide, Protein-tyrosine kinase 2-beta | | Authors: | Han, S. | | Deposit date: | 2012-09-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Identification of novel series of pyrazole and indole-urea based DFG-out PYK2 inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5ZND
 
 | | 8-mer nanotube derived from 24-mer rHuHF nanocage | | Descriptor: | Ferritin heavy chain | | Authors: | Wang, W.M, Wang, L.L, Zang, J.C, Chen, H, Zhao, G.H, Wang, H.F. | | Deposit date: | 2018-04-09 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Selective Elimination of the Key Subunit Interfaces Facilitates Conversion of Native 24-mer Protein Nanocage into 8-mer Nanorings.
J. Am. Chem. Soc., 140, 2018
|
|
6A5F
 
 | |
4HEU
 
 | |
4HF4
 
 | |
6A5G
 
 | |
6J0W
 
 | | Crystal Structure of Yeast Rtt107 and Nse6 | | Descriptor: | Peptide from DNA repair protein KRE29, Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
3GZT
 
 | | VP7 recoated rotavirus DLP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Outer capsid glycoprotein VP7 | | Authors: | Chen, J.Z, Settembre, E.C, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2009-04-07 | | Release date: | 2009-07-14 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GZU
 
 | | VP7 recoated rotavirus DLP | | Descriptor: | Inner capsid protein VP2, Intermediate capsid protein VP6, ZINC ION | | Authors: | Chen, J.Z, Settembre, E.C, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2009-04-07 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6A5H
 
 | |
3S8M
 
 | | The Crystal Structure of FabV | | Descriptor: | Enoyl-ACP Reductase | | Authors: | Li, H, Zhang, X.L, Bi, L.J, He, J, Jiang, T. | | Deposit date: | 2011-05-29 | | Release date: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Determination of the Crystal Structure and Active Residues of FabV, the Enoyl-ACP Reductase from Xanthomonas oryzae.
Plos One, 6, 2011
|
|
4HVA
 
 | |
3HDB
 
 | | Crystal structure of AaHIV, A metalloproteinase from venom of Agkistrodon Acutus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AAHIV, CALCIUM ION, ... | | Authors: | Zhu, Z.Q, Niu, L.W, Teng, M.K. | | Deposit date: | 2009-05-07 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis of the autolysis of AaHIV suggests a novel target recognizing model for ADAM/reprolysin family proteins
Biochem.Biophys.Res.Commun., 386, 2009
|
|
4FYO
 
 | | Crystal structure of spleen tyrosine kinase complexed with N-{(S)-1-[7-(3,4-Dimethoxy-phenylamino)-thiazolo[5,4-d]pyrimidin-5-yl]-pyrrolidin-3-yl}-terephthalamic acid | | Descriptor: | 4-{[(3S)-1-{7-[(3,4-dimethoxyphenyl)amino][1,3]thiazolo[5,4-d]pyrimidin-5-yl}pyrrolidin-3-yl]carbamoyl}benzoic acid, Tyrosine-protein kinase SYK | | Authors: | Kuglstatter, A, Slade, M. | | Deposit date: | 2012-07-05 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rational design of highly selective spleen tyrosine kinase inhibitors.
J.Med.Chem., 55, 2012
|
|
2N31
 
 | |
