4NPT
 
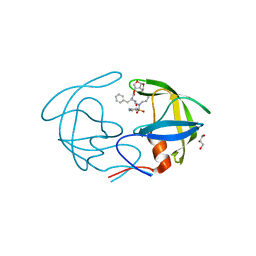 | | Crystal Structure of HIV-1 Protease Multiple Mutant P51 Complexed with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, GLYCEROL, Protease | | Authors: | Zhang, Y, Weber, I.T. | | Deposit date: | 2013-11-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structures of darunavir-resistant HIV-1 protease mutant reveal atypical binding of darunavir to wide open flaps.
Acs Chem.Biol., 9, 2014
|
|
4NPU
 
 | |
5VTM
 
 | |
6A37
 
 | |
8JY0
 
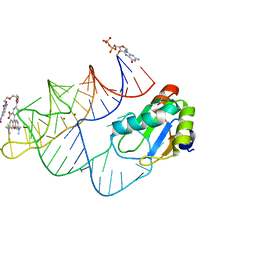 | | Crystal structure of RhoBAST complexed with TMR-DN | | Descriptor: | 2,4-dinitroaniline, 5-aminocarbonyl-2-[3-(dimethylamino)-6-dimethylazaniumylidene-xanthen-9-yl]benzoate, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Y, Xiao, Y, Xu, Z, Fang, X. | | Deposit date: | 2023-07-02 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural mechanisms for binding and activation of a contact-quenched fluorophore by RhoBAST.
Nat Commun, 15, 2024
|
|
7VP1
 
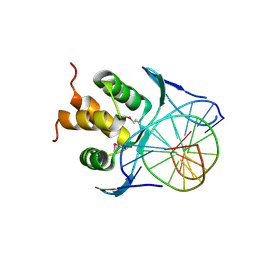 | | Structure of a transcription factor and DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*GP*GP*TP*CP*CP*CP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*GP*GP*GP*AP*CP*CP*AP*CP*A)-3'), Transcription factor TCP10 | | Authors: | Zhang, Y, Xu, Y.P, Wang, B, Su, X.D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural basis for DNA recognition by TCP transcription factors
To Be Published
|
|
7VP2
 
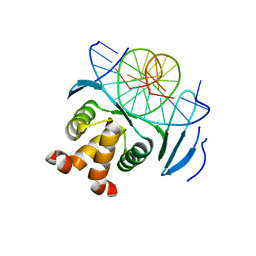 | | Structure of a transcription factor and DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*GP*GP*TP*CP*CP*CP*CP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*GP*GP*GP*GP*AP*CP*CP*AP*CP*A)-3'), Transcription factor TCP10 | | Authors: | Zhang, Y, Xu, Y.P, Wang, B, Su, X.D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for DNA recognition by TCP transcription factors
To Be Published
|
|
7VP4
 
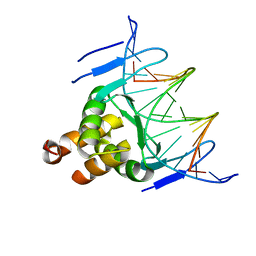 | | Structure of a transcription factor and DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*GP*GP*TP*CP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*GP*GP*GP*GP*AP*CP*CP*AP*CP*A)-3'), Transcription factor TCP10 | | Authors: | Zhang, Y, Xu, Y.P, Wang, B, Su, X.D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural basis for DNA recognition by TCP transcription factors
To Be Published
|
|
7VP7
 
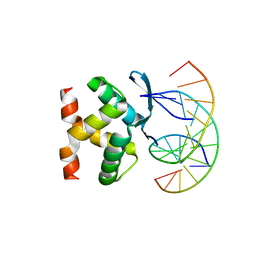 | |
7VP6
 
 | |
7VP5
 
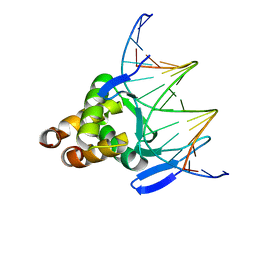 | |
7VP3
 
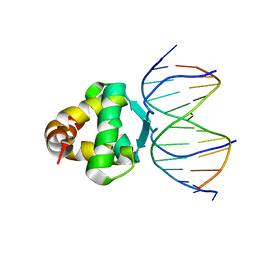 | | Structure of a transcription factor and DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*GP*GP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*GP*GP*AP*CP*CP*CP*AP*C)-3'), Transcription factor TCP15 | | Authors: | Zhang, Y, Xu, Y.P, Wang, B, Su, X.D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural basis for DNA recognition by TCP transcription factors
To Be Published
|
|
9ATN
 
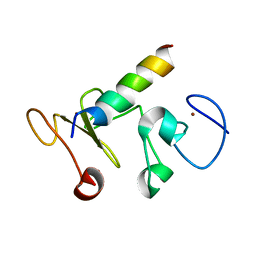 | |
8ISK
 
 | | Pr conformer of Zea mays phytochrome A1 - ZmphyA1-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8ISJ
 
 | | Pr conformer of Arabidopsis thaliana phytochrome A - AtphyA-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8ISI
 
 | | Photochromobilin-free form of Arabidopsis thaliana phytochrome A - apo-AtphyA | | Descriptor: | Phytochrome A | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
6L2G
 
 | | Crystal structure of Aspergillus fumigatus mitochondrial acetyl-CoA acetyltransferase | | Descriptor: | Acetyl-CoA-acetyltransferase, putative | | Authors: | Zhang, Y, Wei, W, Raimi, O.G, Ferenbach, A.T, Fang, W. | | Deposit date: | 2019-10-03 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Aspergillus fumigatus Mitochondrial Acetyl Coenzyme A Acetyltransferase as an Antifungal Target.
Appl.Environ.Microbiol., 86, 2020
|
|
6L2C
 
 | | Crystal structure of Aspergillus fumigatus mitochondrial acetyl-CoA acetyltransferase in complex with CoA | | Descriptor: | Acetyl-CoA-acetyltransferase, putative, COENZYME A | | Authors: | Zhang, Y, Wei, W, Raimi, O.G, Ferenbach, A.T, Fang, W. | | Deposit date: | 2019-10-03 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Aspergillus fumigatus Mitochondrial Acetyl Coenzyme A Acetyltransferase as an Antifungal Target.
Appl.Environ.Microbiol., 86, 2020
|
|
8WGB
 
 | | mGlu2-4 heterodimer bound with Gi | | Descriptor: | (1R,2S)-2-[[3,5-bis(chloranyl)phenyl]carbamoyl]cyclohexane-1-carboxylic acid, GLUTAMIC ACID, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, Y, Liu, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of orientated asymmetry in a mGlu heterodimer
To Be Published
|
|
6LN5
 
 | | CryoEM structure of SERCA2b T1032stop in E1-2Ca2+-AMPPCP (class1) | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6LN9
 
 | | CryoEM structure of SERCA2b T1032stop in E2-BeF3- state (class2) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6LN7
 
 | | CryoEM structure of SERCA2b T1032stop in E1-2Ca2+-AMPPCP (class3) | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6LN8
 
 | | CryoEM structure of SERCA2b T1032stop in E2-BeF3- state (class1) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
6LO8
 
 | | Cryo-EM structure of the TIM22 complex from yeast | | Descriptor: | Mitochondrial import inner membrane translocase subunit TIM10, Mitochondrial import inner membrane translocase subunit TIM12, Mitochondrial import inner membrane translocase subunit TIM18, ... | | Authors: | Zhang, Y, Zhou, X, Wu, X, Li, L. | | Deposit date: | 2020-01-04 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structure of the mitochondrial TIM22 complex from yeast.
Cell Res., 31, 2021
|
|
6LLE
 
 | | CryoEM structure of SERCA2b WT in E1-2Ca2+-AMPPCP state. | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Tsutsumi, A, Watanabe, S, Inaba, K. | | Deposit date: | 2019-12-23 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail.
Sci Adv, 6, 2020
|
|
