1TPV
 
 | | S96P CHANGE IS A SECOND-SITE SUPPRESSOR FOR H95N SLUGGISH MUTANT TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Stock, A.M, Narayana, N, Xuong, Ng.H, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-11-07 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the H95N lesion by the secondary S96P mutation in triosephosphate isomerase.
Biochemistry, 35, 1996
|
|
1TPW
 
 | | TRIOSEPHOSPHATE ISOMERASE DRINKS WATER TO KEEP HEALTHY | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Stock, A.M, Narayana, N, Xuong, Ng.H, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-11-07 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of water in the catalytic efficiency of triosephosphate isomerase.
Biochemistry, 38, 1999
|
|
1B4X
 
 | | ASPARTATE AMINOTRANSFERASE FROM E. COLI, C191S MUTATION, WITH BOUND MALEATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jeffery, C.J, Gloss, L.M, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-12-30 | | Release date: | 2000-10-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The role of residues outside the active site: structural basis for function of C191 mutants of Escherichia coli aspartate aminotransferase.
Protein Eng., 13, 2000
|
|
1QIR
 
 | | ASPARTATE AMINOTRANSFERASE FROM ESCHERICHIA COLI, C191Y MUTATION, WITH BOUND MALEATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jeffery, C.J, Gloss, L.M, Petsko, G.A, Ringe, D. | | Deposit date: | 1999-06-15 | | Release date: | 2000-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Role of Residues Outside the Active Site in Catalysis: Structural Basis for Function of C191 Mutants of E. Coli Aspartate Aminotransferase
Protein Eng., 13, 2000
|
|
1QIT
 
 | | ASPARTATE AMINOTRANSFERASE FROM ESCHERICHIA COLI, C191W MUTATION, WITH BOUND MALEATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jeffery, C.J, Gloss, L.M, Petsko, G.A, Ringe, D. | | Deposit date: | 1999-06-15 | | Release date: | 2000-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Role of Residues Outside the Active Site in Catalysis: Structural Basis for Function of C191 Mutants of E. Coli Aspartate Aminotransferase
Protein Eng., 13, 2000
|
|
1QIS
 
 | | ASPARTATE AMINOTRANSFERASE FROM ESCHERICHIA COLI, C191F MUTATION, WITH BOUND MALEATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jeffery, C.J, Gloss, L.M, Petsko, G.A, Ringe, D. | | Deposit date: | 1999-06-15 | | Release date: | 2000-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Role of Residues Outside the Active Site in Catalysis: Structural Basis for Function of C191 Mutants of E. Coli Aspartate Aminotransferase
Protein Eng., 13, 2000
|
|
1OT5
 
 | | The 2.4 Angstrom Crystal Structure of Kex2 in complex with a peptidyl-boronic acid inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ac-Ala-Lys-boroArg N-acetylated boronic acid peptide inhibitor, ... | | Authors: | Holyoak, T, Wilson, M.A, Fenn, T.D, Kettner, C.A, Petsko, G.A, Fuller, R.S, Ringe, D. | | Deposit date: | 2003-03-21 | | Release date: | 2003-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 A Resolution Crystal Structure of the Prototypical Hormone-Processing Protease Kex2 in Complex with an Ala-Lys-Arg Boronic Acid Inhibitor
Biochemistry, 42, 2003
|
|
1AZ2
 
 | | CITRATE BOUND, C298A/W219Y MUTANT HUMAN ALDOSE REDUCTASE | | Descriptor: | ALDOSE REDUCTASE, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H, Bohren, K.M, Ringe, D, Petsko, G.A, Gabbay, K.H. | | Deposit date: | 1997-11-24 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The alrestatin double-decker: binding of two inhibitor molecules to human aldose reductase reveals a new specificity determinant.
Biochemistry, 36, 1997
|
|
1MR3
 
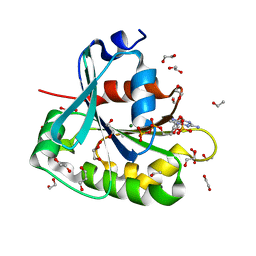 | | Saccharomyces cerevisiae ADP-ribosylation Factor 2 (ScArf2) complexed with GDP-3'P at 1.6A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ADP-ribosylation factor 2, ... | | Authors: | Amor, J.-C, Horton, J.R, Zhu, X, Wang, Y, Sullards, C, Ringe, D, Cheng, X, Kahn, R.A. | | Deposit date: | 2002-09-17 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of yeast ARF2 and ARL1: distinct roles for the N terminus in the structure and function of ARF family GTPases.
J.Biol.Chem., 276, 2001
|
|
1P92
 
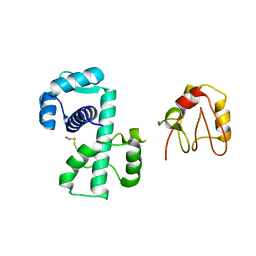 | | Crystal Structure of (H79A)DtxR | | Descriptor: | BETA-MERCAPTOETHANOL, Diphtheria toxin repressor | | Authors: | D'Aquino, J.A, Ringe, D. | | Deposit date: | 2003-05-08 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Determinants of the SRC homology domain 3-like fold.
J.Bacteriol., 185, 2003
|
|
1P5F
 
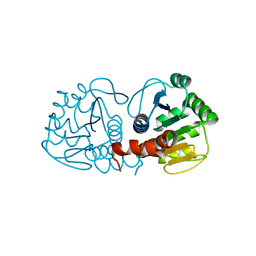 | | Crystal Structure of Human DJ-1 | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Wilson, M.A, Collins, J.L, Hod, Y, Ringe, D, Petsko, G.A. | | Deposit date: | 2003-04-26 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson's disease
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1M54
 
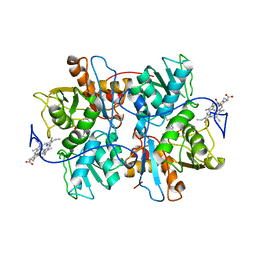 | | CYSTATHIONINE-BETA SYNTHASE: REDUCED VICINAL THIOLS | | Descriptor: | CYSTATHIONINE BETA-SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Taoka, S, Lepore, B.W, Kabil, O, Ojha, S, Ringe, D, Banerjee, R. | | Deposit date: | 2002-07-08 | | Release date: | 2002-08-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | HUMAN CYSTATHIONINE BETA-SYNTHASE IS A HEME SENSOR PROTEIN. EVIDENCE THAT THE
REDOX SENSOR IS HEME AND NOT THE VICINAL CYSTEINES IN THE CXXC MOTIF SEEN IN THE CRYSTAL STRUCTURE OF THE TRUNCATED ENZYME
BIOCHEMISTRY, 41, 2002
|
|
1NIU
 
 | | ALANINE RACEMASE WITH BOUND INHIBITOR DERIVED FROM L-CYCLOSERINE | | Descriptor: | Alanine Racemase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Fenn, T.D, Stamper, G.F, Morollo, A.A, Ringe, D. | | Deposit date: | 2002-12-26 | | Release date: | 2003-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A side reaction of alanine racemase: transamination of cycloserine.
Biochemistry, 42, 2003
|
|
1Q0X
 
 | | Anti-morphine Antibody 9B1 Unliganded Form | | Descriptor: | Fab 9B1, heavy chain, light chain, ... | | Authors: | Pozharski, E, Wilson, M.A, Hewagama, A, Shanafelt, A.B, Petsko, G, Ringe, D. | | Deposit date: | 2003-07-17 | | Release date: | 2004-04-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Anchoring a cationic ligand: the structure of the Fab fragment of the anti-morphine antibody 9B1 and its complex with morphine
J.Mol.Biol., 337, 2004
|
|
1AZ1
 
 | | ALRESTATIN BOUND TO C298A/W219Y MUTANT HUMAN ALDOSE REDUCTASE | | Descriptor: | ALDOSE REDUCTASE, ALRESTATIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Bohren, K.M, Petsko, G.A, Ringe, D, Gabbay, K.H. | | Deposit date: | 1997-11-24 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The alrestatin double-decker: binding of two inhibitor molecules to human aldose reductase reveals a new specificity determinant.
Biochemistry, 36, 1997
|
|
1Q0Y
 
 | | Anti-Morphine Antibody 9B1 Complexed with Morphine | | Descriptor: | (7R,7AS,12BS)-3-METHYL-2,3,4,4A,7,7A-HEXAHYDRO-1H-4,12-METHANO[1]BENZOFURO[3,2-E]ISOQUINOLINE-7,9-DIOL, Fab 9B1, Heavy chain, ... | | Authors: | Pozharski, E, Wilson, M.A, Hewagama, A, Shanafelt, A.B, Petsko, G, Ringe, D. | | Deposit date: | 2003-07-17 | | Release date: | 2004-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Anchoring a cationic ligand: the structure of the Fab fragment of the anti-morphine antibody 9B1 and its complex with morphine
J.Mol.Biol., 337, 2004
|
|
1BRM
 
 | | ASPARTATE BETA-SEMIALDEHYDE DEHYDROGENASE FROM ESCHERICHIA COLI | | Descriptor: | ASPARTATE-SEMIALDEHYDE DEHYDROGENASE | | Authors: | Hadfield, A.T, Kryger, G, Ouyang, J, Ringe, D, Petsko, G.A, Viola, R.E. | | Deposit date: | 1998-08-24 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of aspartate-beta-semialdehyde dehydrogenase from Escherichia coli, a key enzyme in the aspartate family of amino acid biosynthesis.
J.Mol.Biol., 289, 1999
|
|
1MOZ
 
 | | ADP-ribosylation factor-like 1 (ARL1) from Saccharomyces cerevisiae | | Descriptor: | ADP-ribosylation factor-like protein 1, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Amor, J.C, Horton, J.R, Zhu, X, Wang, Y, Sullards, C, Ringe, D, Cheng, X, Kahn, R.A. | | Deposit date: | 2002-09-10 | | Release date: | 2002-10-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structures of Yeast ARF2 and ARL1:
DISTINCT ROLES FOR THE N TERMINUS IN THE STRUCTURE
AND FUNCTION OF ARF FAMILY GTPases
J.Biol.Chem., 276, 2001
|
|
1Q72
 
 | | Anti-Cocaine Antibody M82G2 Complexed with Cocaine | | Descriptor: | COCAINE, Fab M82G2, Heavy chain, ... | | Authors: | Pozharski, E, Moulin, A, Hewagama, A, Shanafelt, A.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2003-08-15 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Diversity in hapten recognition: structural study of an anti-cocaine antibody M82G2.
J.Mol.Biol., 349, 2005
|
|
1BKH
 
 | | MUCONATE LACTONIZING ENZYME FROM PSEUDOMONAS PUTIDA | | Descriptor: | MUCONATE LACTONIZING ENZYME | | Authors: | Hasson, M.S, Schlichting, I, Moulai, J, Taylor, K, Barrett, W, Kenyon, G.L, Babbitt, P.C, Gerlt, J.A, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-07-07 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of an enzyme active site: the structure of a new crystal form of muconate lactonizing enzyme compared with mandelate racemase and enolase.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1CM7
 
 | | 3-ISOPROPYLMALATE DEHYDROGENASE FROM ESCHERICHIA COLI | | Descriptor: | PROTEIN (3-ISOPROPYLMALATE DEHYDROGENASE) | | Authors: | Wallon, G, Kryger, G, Lovett, S.T, Oshima, T, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-05-17 | | Release date: | 1999-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of Escherichia coli and Salmonella typhimurium 3-isopropylmalate dehydrogenase and comparison with their thermophilic counterpart from Thermus thermophilus.
J.Mol.Biol., 266, 1997
|
|
1MUW
 
 | |
1CP6
 
 | | 1-BUTANEBORONIC ACID BINDING TO AEROMONAS PROTEOLYTICA AMINOPEPTIDASE | | Descriptor: | 1-BUTANE BORONIC ACID, PROTEIN (AMINOPEPTIDASE), ZINC ION | | Authors: | Depaola, C.C, Bennett, B, Holz, R.C, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-06-08 | | Release date: | 1999-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1-Butaneboronic acid binding to Aeromonas proteolytica aminopeptidase: a case of arrested development.
Biochemistry, 38, 1999
|
|
1ESB
 
 | | DIRECT STRUCTURE OBSERVATION OF AN ACYL-ENZYME INTERMEDIATE IN THE HYDROLYSIS OF AN ESTER SUBSTRATE BY ELASTASE | | Descriptor: | CALCIUM ION, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, PORCINE PANCREATIC ELASTASE, ... | | Authors: | Ding, X, Rasmussen, B, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-04 | | Release date: | 1994-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Direct structural observation of an acyl-enzyme intermediate in the hydrolysis of an ester substrate by elastase.
Biochemistry, 33, 1994
|
|
2SFP
 
 | | ALANINE RACEMASE WITH BOUND PROPIONATE INHIBITOR | | Descriptor: | PROPANOIC ACID, PROTEIN (ALANINE RACEMASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Morollo, A.A, Petsko, G.A, Ringe, D. | | Deposit date: | 1999-02-16 | | Release date: | 1999-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Michaelis complex analogue: propionate binds in the substrate carboxylate site of alanine racemase.
Biochemistry, 38, 1999
|
|
