6Z6H
 
 | | HDAC-DC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (8.55 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6F
 
 | | HDAC-PC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6P
 
 | | HDAC-PC-Nuc | | Descriptor: | DNA (145-MER), HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6O
 
 | | HDAC-TC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
4OWI
 
 | | peptide structure | | Descriptor: | p53LZ2 | | Authors: | Lee, J.-H. | | Deposit date: | 2014-02-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.202 Å) | | Cite: | Protein grafting of p53TAD onto a leucine zipper scaffold generates a potent HDM dual inhibitor.
Nat Commun, 5, 2014
|
|
5Y3D
 
 | |
4LCT
 
 | | Crystal Structure and Versatile Functional Roles of the COP9 Signalosome Subunit 1 | | Descriptor: | COP9 signalosome complex subunit 1, SULFATE ION | | Authors: | Lee, J.-H, Yi, L, Li, J, Schweitzer, K, Borgmann, M, Naumann, M, Wu, H. | | Deposit date: | 2013-06-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and versatile functional roles of the COP9 signalosome subunit 1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1CQ0
 
 | | SOLUTION STRUCTURE OF A HUMAN HYPOCRETIN-2/OREXIN-B'SOLUTION STRUCTURE OF A HUMAN HYPOCRETIN-2/OREXIN-B ' | | Descriptor: | PROTEIN (NEW HYPOTHALAMIC NEUROPEPTIDE/OREXIN-B28) | | Authors: | Lee, K.-H, Bang, E.J, Chae, K.-J, Lee, D.W, Lee, W. | | Deposit date: | 1999-08-04 | | Release date: | 2000-01-10 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a new hypothalamic neuropeptide, human hypocretin-2/orexin-B.
Eur.J.Biochem., 266, 1999
|
|
1X3Z
 
 | | Structure of a peptide:N-glycanase-Rad23 complex | | Descriptor: | UV excision repair protein RAD23, ZINC ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, ... | | Authors: | Lee, J.-H, Choi, J.M, Lee, C, Yi, K.J, Cho, Y. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a peptide:N-glycanase-Rad23 complex: insight into the deglycosylation for denatured glycoproteins.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1X3W
 
 | | Structure of a peptide:N-glycanase-Rad23 complex | | Descriptor: | UV excision repair protein RAD23, ZINC ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, ... | | Authors: | Lee, J.-H, Choi, J.M, Lee, C, Yi, K.J, Cho, Y. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a peptide:N-glycanase-Rad23 complex: insight into the deglycosylation for denatured glycoproteins.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1CFL
 
 | | DNA DECAMER DUPLEX CONTAINING T5-T6 PHOTOADDUCT | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*(64T)P*TP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*TP*GP*CP*G)-3') | | Authors: | Lee, J.-H, Hwang, G.-S, Choi, B.-S. | | Deposit date: | 1999-03-19 | | Release date: | 1999-05-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA decamer duplex containing the stable 3' T.G base pair of the pyrimidine(6-4)pyrimidone photoproduct [(6-4) adduct]: implications for the highly specific 3' T --> C transition of the (6-4) adduct.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6WWX
 
 | |
6WV2
 
 | |
6WXA
 
 | | Crystal structure of truncated Streptococcal bacteriophage hyaluronidase complexed with unsaturated hyaluronan hexa-saccharides | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Deivanayagam, C, Schormann, N. | | Deposit date: | 2020-05-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Streptococcal Bacteriophage Hyaluronidase:
Presence of a Prokaryotic Collagen and Elucidation of Catalytic Mechanism
To Be Published
|
|
6X3M
 
 | | Crystal structure of full-length Streptococcal bacteriophage hyaluronidase in complex with unsaturated hyaluronan octa-saccharides | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hyaluronoglucosaminidase | | Authors: | Deivanayagam, C, Schormann, N. | | Deposit date: | 2020-05-21 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.581 Å) | | Cite: | Crystal Structure of Streptococcal Bacteriophage Hyaluronidase:
Presence of a Prokaryotic Collagen and Elucidation of Catalytic Mechanism
To Be Published
|
|
3OU6
 
 | | DhpI-SAM complex | | Descriptor: | S-ADENOSYLMETHIONINE, SAM-dependent methyltransferase, SULFATE ION | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2010-09-14 | | Release date: | 2010-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OU7
 
 | | DhpI-SAM-HEP complex | | Descriptor: | (2-hydroxyethyl)phosphonic acid, S-ADENOSYLMETHIONINE, SAM-dependent methyltransferase, ... | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2010-09-14 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OU2
 
 | | DhpI-SAH complex structure | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2010-09-14 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5H22
 
 | | Hsp90 alpha N-terminal domain in complex with an inhibitor | | Descriptor: | 4-chloranyl-7-[(4-methoxy-3,5-dimethyl-pyridin-2-yl)methyl]-5-(phenylmethyl)pyrrolo[2,3-d]pyrimidin-2-amine, Hsp90aa1 protein | | Authors: | Kim, E.E, Shin, S.C, Keum, G.C. | | Deposit date: | 2016-10-13 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Synthesis and in vitro antiproliferative activity of C5-benzyl substituted 2-amino-pyrrolo[2,3-d]pyrimidines as potent Hsp90 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7YLK
 
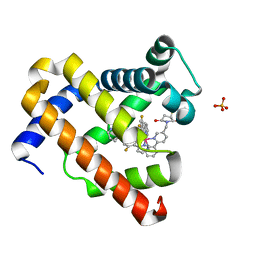 | | Myoglobin containing Ir complex | | Descriptor: | Myoglobin, SULFATE ION, delta-{1-([2,2'-bipyridin]-5-ylmethyl)pyrrolidine-2,5-dione}bis[2-(2,4-difluorophenyl)pyridine)]iridium(III), ... | | Authors: | Lee, J.H, Song, W.J. | | Deposit date: | 2022-07-26 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Photocatalytic C-O Coupling Enzymes That Operate via Intramolecular Electron Transfer.
J.Am.Chem.Soc., 145, 2023
|
|
6IKZ
 
 | |
6IKX
 
 | |
6K8D
 
 | |
6KNJ
 
 | |
6KNL
 
 | |
