2Z4W
 
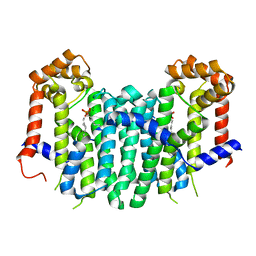 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with magnesium and BPH-749 | | Descriptor: | Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION, [(6E,11E)-2,6,12,16-tetramethylheptadeca-2,6,11,15-tetraene-9,9-diyl]bis(phosphonic acid) | | Authors: | Chen, C.K.-M, Guo, R.T, Hudock, M, Cao, R, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-06-26 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inhibition of geranylgeranyl diphosphate synthase by bisphosphonates: a crystallographic and computational investigation
J.Med.Chem., 51, 2008
|
|
8WGZ
 
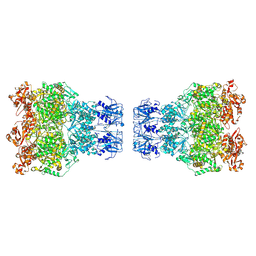 | | MPOX E5 double hexamer ssDNA bound conformation | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*C)-3'), Uncoating factor OPG117 | | Authors: | Zhang, Z, Dong, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Essential and multifunctional mpox virus E5 helicase-primase in double and single hexamer.
Sci Adv, 10, 2024
|
|
8WH0
 
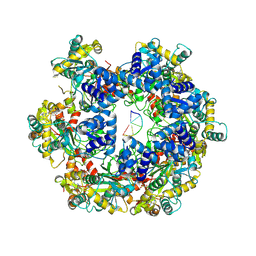 | | MPOX E5 hexamer ssDNA and AMP-PNP bound conformation | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*C)-3'), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhang, Z, Dong, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Essential and multifunctional mpox virus E5 helicase-primase in double and single hexamer.
Sci Adv, 10, 2024
|
|
8WGY
 
 | |
2Z4Z
 
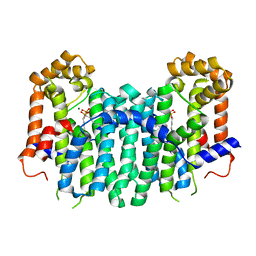 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with magnesium and BPH-sc01 | | Descriptor: | (6E,11E)-HEPTADECA-6,11-DIENE-9,9-DIYLBIS(PHOSPHONIC ACID), Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Chen, C.K.-M, Guo, R.T, Hudock, M, Cao, R, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-06-26 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Inhibition of geranylgeranyl diphosphate synthase by bisphosphonates: a crystallographic and computational investigation
J.Med.Chem., 51, 2008
|
|
2Z50
 
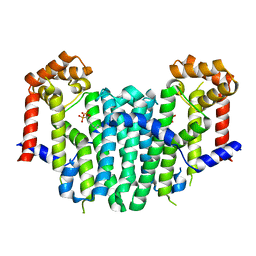 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with BPH-28 | | Descriptor: | (1-HYDROXYHEPTANE-1,1-DIYL)BIS(PHOSPHONIC ACID), Geranylgeranyl pyrophosphate synthetase | | Authors: | Guo, R.T, Chen, C.K.-M, Hudock, M, Cao, R, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-06-26 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Inhibition of geranylgeranyl diphosphate synthase by bisphosphonates: a crystallographic and computational investigation
J.Med.Chem., 51, 2008
|
|
5Y6G
 
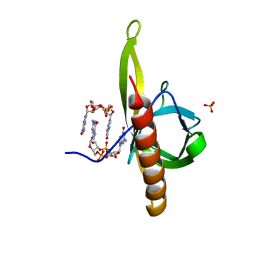 | | PilZ domain with c-di-GMP of YcgR from Escherichia coli | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Flagellar brake protein YcgR, SULFATE ION | | Authors: | Hou, Y.J, Wang, D.C, Li, D.F. | | Deposit date: | 2017-08-11 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility inEscherichia coli.
J.Biol.Chem., 295, 2020
|
|
5Y6H
 
 | |
5Z2L
 
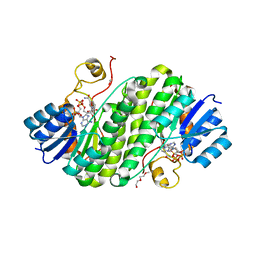 | | Crystal structure of BdcA in complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Cyclic-di-GMP-binding biofilm dispersal mediator protein, ... | | Authors: | Yang, W.S, Hou, Y.J, Li, D.F, Wang, D.C. | | Deposit date: | 2018-01-03 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A potential substrate binding pocket of BdcA plays a critical role in NADPH recognition and biofilm dispersal
Biochem. Biophys. Res. Commun., 497, 2018
|
|
2Z78
 
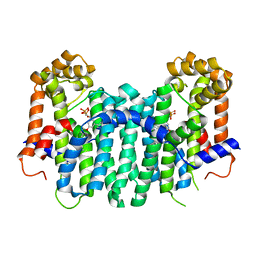 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with BPH-806 | | Descriptor: | 3-(decyloxy)-5-(3,5-difluorophenyl)-1-(2,2-diphosphonoethyl)pyridinium, Geranylgeranyl pyrophosphate synthetase | | Authors: | Chen, C.K.-M, Guo, R.T, Hudock, M, Cao, R, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-08-16 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of geranylgeranyl diphosphate synthase by bisphosphonates: a crystallographic and computational investigation
J.Med.Chem., 51, 2008
|
|
2Z4Y
 
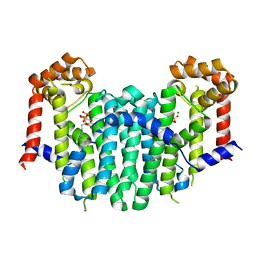 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with magnesium and BPH-252 | | Descriptor: | (1-HYDROXYNONANE-1,1-DIYL)BIS(PHOSPHONIC ACID), Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Chen, C.K.-M, Guo, R.T, Hudock, M, Cao, R, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-06-26 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of geranylgeranyl diphosphate synthase by bisphosphonates: a crystallographic and computational investigation
J.Med.Chem., 51, 2008
|
|
3ADZ
 
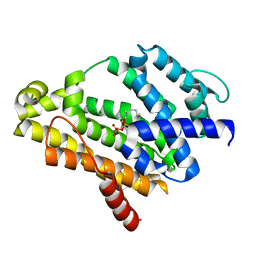 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with intermediate PSPP | | Descriptor: | Dehydrosqualene synthase, MAGNESIUM ION, {(1R,2R,3R)-2-[(3E)-4,8-dimethylnona-3,7-dien-1-yl]-2-methyl-3-[(1E,5E)-2,6,10-trimethylundeca-1,5,9-trien-1-yl]cyclopropyl}methyl trihydrogen diphosphate | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J, Oldfield, E. | | Deposit date: | 2010-01-31 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Mechanism of action and inhibition of dehydrosqualene synthase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3AE0
 
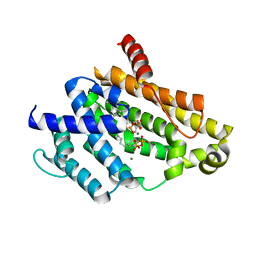 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with geranylgeranyl thiopyrophosphate | | Descriptor: | Dehydrosqualene synthase, MAGNESIUM ION, phosphonooxy-[(10E)-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraenyl]sulfanyl-phosphinic acid | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J, Oldfield, E. | | Deposit date: | 2010-01-31 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mechanism of action and inhibition of dehydrosqualene synthase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8XVJ
 
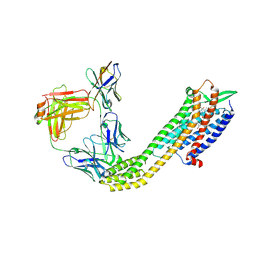 | | Cryo-EM structure of ETAR bound with Macitentan | | Descriptor: | 6-[2-(5-bromanylpyrimidin-2-yl)oxyethoxy]-5-(4-bromophenyl)-~{N}-(propylsulfamoyl)pyrimidin-4-amine, Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVK
 
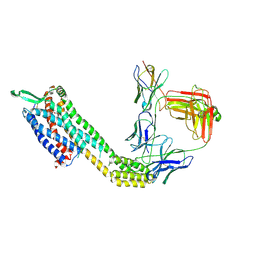 | | Cryo-EM structure of ETAR bound with Ambrisentan | | Descriptor: | (2~{S})-2-(4,6-dimethylpyrimidin-2-yl)oxy-3-methoxy-3,3-diphenyl-propanoic acid, Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVL
 
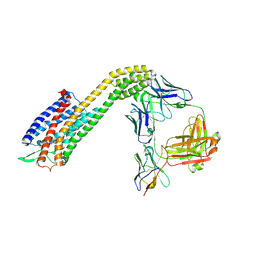 | | Cryo-EM structure of ETAR bound with Zibotentan | | Descriptor: | Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, anti-BRIL Fab Light chain, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
1G69
 
 | | THIAMIN PHOSPHATE SYNTHASE | | Descriptor: | 2-METHYL-5-METHYLENE-5H-PYRIMIDIN-4-YLIDENEAMINE, 4-METHYL-5-HYDROXYETHYLTHIAZOLE PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Peapus, D.H, Chiu, H.-J, Campobasso, N, Reddick, J.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-11-03 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes of thiamin phosphate synthase.
Biochemistry, 40, 2001
|
|
8XVI
 
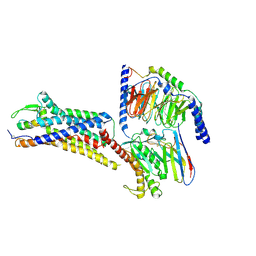 | | Cryo-EM structure of ETAR bound with Endothelin1 | | Descriptor: | Endoglucanase H,Endothelin-1 receptor, Endothelin-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVH
 
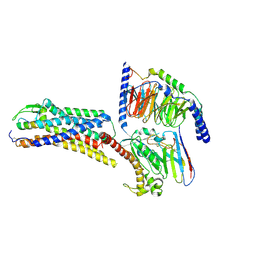 | | Cryo-EM structure of ETBR bound with Endothelin1 | | Descriptor: | Endothelin-1, Exo-alpha-sialidase,Endothelin receptor type B, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVE
 
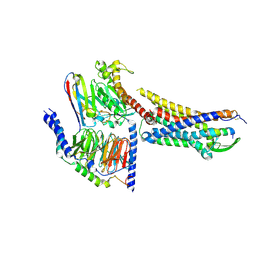 | | Cryo-EM structure of ETBR bound with BQ3020 | | Descriptor: | BQ3020, Exo-alpha-sialidase,Endothelin receptor type B, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
1G4P
 
 | | THIAMIN PHOSPHATE SYNTHASE | | Descriptor: | 4-AMINO-2-TRIFLUOROMETHYL-5-HYDROXYMETHYLPYRIMIDINE PYROPHOSPHATE, MAGNESIUM ION, THIAMIN PHOSPHATE SYNTHASE | | Authors: | Peapus, D.H, Chiu, H.-J, Campobasso, N, Reddick, J.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-10-27 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural characterization of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes of thiamin phosphate synthase.
Biochemistry, 40, 2001
|
|
6IRA
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 7.8 | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
7YKC
 
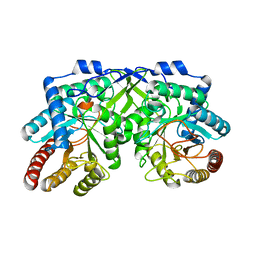 | |
6IRH
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class III | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRF
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class I | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
