2KMM
 
 | | Solution NMR structure of the TGS domain of PG1808 from Porphyromonas gingivalis. Northeast Structural Genomics Consortium Target PgR122A (418-481) | | Descriptor: | Guanosine-3',5'-bis(Diphosphate) 3'-pyrophosphohydrolase | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Lee, D, Ciccosanti, C, Hamilton, K, Nair, R, Rost, B, Acton, T.B, Xiao, R, Swapna, G, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-07-31 | | Release date: | 2009-11-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the TGS domain of PG1808 from
Porphyromonas gingivalis. Northeast Structural Genomics Consortium Target
PgR122A (418-481)
To be Published
|
|
2KJ7
 
 | | Three-Dimensional NMR Structure of Rat Islet Amyloid Polypeptide in DPC micelles | | Descriptor: | Islet amyloid polypeptide | | Authors: | Nanga, R, Brender, J.R, Xu, J, Hartman, K, Subramanian, V, Ramamoorthy, A. | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-23 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and orientation of rat islet amyloid polypeptide protein in a membrane environment by solution NMR spectroscopy.
J.Am.Chem.Soc., 131, 2009
|
|
2KKO
 
 | | Solution NMR structure of the homodimeric winged helix-turn-helix DNA-binding domain (fragment 1-100) Mb0332 from Mycobacterium bovis, a possible ArsR-family transcriptional regulator. Northeast Structural Genomics Consortium Target MbR242E. | | Descriptor: | POSSIBLE TRANSCRIPTIONAL REGULATORY PROTEIN (POSSIBLY ARSR-FAMILY) | | Authors: | Ramelot, T.A, Cort, J.R, Wang, D, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Swapna, G, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-26 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the homodimeric winged helix-turn-helix DNA-binding domain (fragment 1-100) Mb0332 from Mycobacterium bovis, a possible ArsR-family transcriptional regulator. Northeast Structural Genomics Consortium Target MbR242E.
To be Published
|
|
2JQO
 
 | | NMR solution structure of Bacillus subtilis YobA 21-120: Northeast Structural Genomics Consortium target SR547 | | Descriptor: | Hypothetical protein yobA | | Authors: | Cort, J.R, Ramelot, T.A, Chen, C.X, Jang, M, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of Bacillus subtilis YobA 21-120: Northeast Structural Genomics Consortium target SR547
To be Published
|
|
2JIZ
 
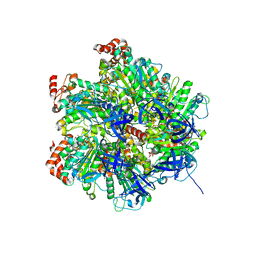 | | The Structure of F1-ATPase inhibited by resveratrol. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE GAMMA CHAIN, ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, ... | | Authors: | Gledhill, J.R, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Inhibition of Bovine F1-ATPase by Resveratrol and Related Polyphenols.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2KF2
 
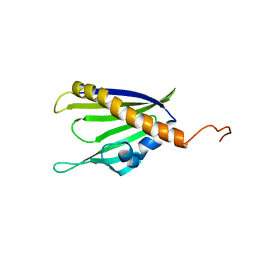 | | Solution NMR structure of of Streptomyces coelicolor polyketide cyclase SCO5315. Northeast Structural Genomics Consortium target RR365 | | Descriptor: | Putative polyketide cyclase | | Authors: | Cort, J.R, Ramelot, T.A, Ding, K, Wang, H, Jiang, M, Maglaqui, M, Xiao, R, Nair, R, Baran, M.C, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-11 | | Release date: | 2009-05-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Streptomyces coelicolor polyketide cyclase SCO5315
To be Published
|
|
2K4Z
 
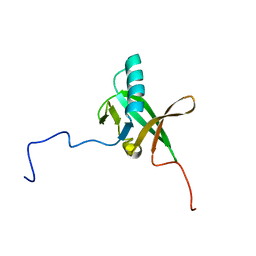 | | Solution NMR Structure of Allochromatium vinosum DsrR: Northeast Structural Genomics Consortium Target OP5 | | Descriptor: | DsrR | | Authors: | Cort, J.R, Dahl, C, Grimm, F, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-22 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the IscA-like Protein DsrR Involved in Sulfur Oxidation in Allochromatium vinosum
To be Published
|
|
2KJR
 
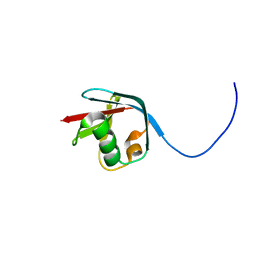 | | Solution NMR structure of the N-terminal Ubiquitin-like Domain from Tubulin-binding Cofactor B, CG11242, from Drosophila melanogaster. Northeast Structural Genomics Consortium Target FR629A (residues 8-92) | | Descriptor: | CG11242 | | Authors: | Ramelot, T.A, Cort, J.R, Shastry, R, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Swapna, G, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-08 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the N-terminal Ubiquitin-like Domain from
Tubulin-binding Cofactor B, CG11242, from Drosophila melanogaster. Northeast
Structural Genomics Consortium Target FR629A (residues 8-92)
To be Published
|
|
2HAC
 
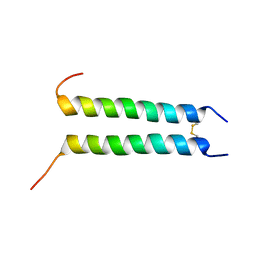 | | Structure of Zeta-Zeta Transmembrane Dimer | | Descriptor: | T-cell surface glycoprotein CD3 zeta chain | | Authors: | Chou, J.J, Wucherpfennig, K.W, Schnell, J.R, Call, M.E. | | Deposit date: | 2006-06-12 | | Release date: | 2006-10-31 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | The structure of the zetazeta transmembrane dimer reveals features essential for its assembly with the T cell receptor.
Cell(Cambridge,Mass.), 127, 2006
|
|
2H6D
 
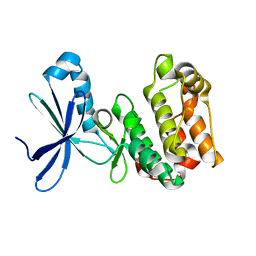 | | Protein Kinase Domain of the Human 5'-AMP-activated protein kinase catalytic subunit alpha-2 (AMPK alpha-2 chain) | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2 | | Authors: | Littler, D.R, Walker, J.R, Wybenga-Groot, L, Newman, E.M, Butler-Cole, C, Mackenzie, F, Finerty, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-31 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A conserved mechanism of autoinhibition for the AMPK kinase domain: ATP-binding site and catalytic loop refolding as a means of regulation.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2HE9
 
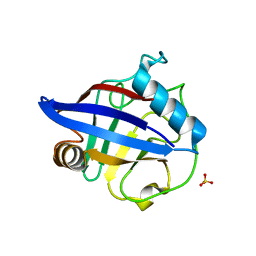 | | Structure of the peptidylprolyl isomerase domain of the human NK-tumour recognition protein | | Descriptor: | NK-tumor recognition protein, SULFATE ION | | Authors: | Walker, J.R, Davis, T, Newman, E.M, MacKenzie, F, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases.
PLoS Biol., 8, 2010
|
|
2HEQ
 
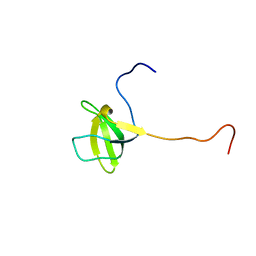 | | NMR Structure of Bacillus subtilis protein YorP, Northeast Structural Genomics Target SR399. | | Descriptor: | YorP protein | | Authors: | Ramelot, T.A, Cort, J.R, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.M, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-21 | | Release date: | 2006-08-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Bacillus subtilis protein YorP, Northeast Structural Genomics Target SR399.
To be Published
|
|
2J98
 
 | | Human coronavirus 229E non structural protein 9 cys69ala mutant (Nsp9) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, REPLICASE POLYPROTEIN 1AB | | Authors: | Ponnusamy, R, Mesters, J.R, Moll, R, Hilgenfeld, R. | | Deposit date: | 2006-11-03 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Variable Oligomerization Modes in Coronavirus Non-Structural Protein 9.
J.Mol.Biol., 383, 2008
|
|
2J97
 
 | | Human coronavirus 229E non structural protein 9 (Nsp9) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, REPLICASE POLYPROTEIN 1AB, SULFATE ION | | Authors: | Ponnusamy, R, Mesters, J.R, Moll, R, Hilgenfeld, R. | | Deposit date: | 2006-11-03 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Variable Oligomerization Modes in Coronavirus Non-Structural Protein 9.
J.Mol.Biol., 383, 2008
|
|
2IRW
 
 | | Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) with NADP and Adamantane Ether Inhibitor | | Descriptor: | (1S,3R,4S,5S,7S)-4-{[2-(4-METHOXYPHENOXY)-2-METHYLPROPANOYL]AMINO}ADAMANTANE-1-CARBOXAMIDE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Longenecker, K.L, Patel, J.R, Russell, J, Qin, W. | | Deposit date: | 2006-10-16 | | Release date: | 2007-01-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of adamantane ethers as inhibitors of 11beta-HSD-1: Synthesis and biological evaluation.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2KNJ
 
 | | NMR structure of microplusin a antimicrobial peptide from Rhipicephalus (Boophilus) microplus | | Descriptor: | Microplusin preprotein | | Authors: | Pires, J.R, Rezende, C.A, Silva, F.D, Daffre, S. | | Deposit date: | 2009-08-26 | | Release date: | 2009-10-13 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and mode of action of microplusin, a copper II-chelating antimicrobial peptide from the cattle tick Rhipicephalus (Boophilus) microplus.
J.Biol.Chem., 284, 2009
|
|
2KPU
 
 | | NMR Structure of YbbR family protein Dhaf_0833 (residues 32-118) from Desulfitobacterium hafniense DCB-2: Northeast Structural Genomics Consortium target DhR29B | | Descriptor: | YbbR family protein | | Authors: | Cort, J.R, Ramelot, T.A, Yang, Y, Belote, R.L, Ciccosanti, C, Haleema, J, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structures of domains I and IV from YbbR are representative of a widely distributed protein family.
Protein Sci., 20, 2011
|
|
2KON
 
 | | NMR solution structure of CV_2116 from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CvT4(1-82) | | Descriptor: | Uncharacterized protein | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Garcia, M, Yee, A, Arrowsmith, C.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-24 | | Release date: | 2009-10-13 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of CV_2116 from Chromobacterium violaceum.Northeast Structural Genomics Consortium Target CvT4(1-82)
To be Published
|
|
2K4N
 
 | | NMR structure of protein PF0246 from Pyrococcus furiosus: target PfR75 from the Northeast Structural Genomics Consortium | | Descriptor: | Protein PF0246 | | Authors: | Cort, J.R, Ho, C.K, Shetty, K, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-13 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution-State NMR Structure of protein PF0246 from Pyrococcus Furiosis
To be Published
|
|
2KPI
 
 | | Solution NMR structure of Streptomyces coelicolor SCO3027 modeled with Zn+2 bound, Northeast Structural Genomics Consortium Target RR58 | | Descriptor: | Uncharacterized protein SCO3027, ZINC ION | | Authors: | Ramelot, T.A, Cort, J.R, Garcia, M, Yee, A, Arrowmith, C.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Streptomyces coelicolor SCO3027 modeled with Zn+2 bound, Northeast Structural Genomics Consortium Target RR58
To be Published
|
|
2KS0
 
 | | Solution NMR structure of the Q251Q8_DESHY(21-82) protein from Desulfitobacterium Hafniense, Northeast Structural Genomics Consortium Target DhR8C | | Descriptor: | Uncharacterized protein | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Wang, H, Ciccosanti, C, Foote, E.L, Jiang, M, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-23 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Combining NMR and EPR methods for homodimer protein structure determination.
J.Am.Chem.Soc., 132, 2010
|
|
2K5T
 
 | | Solution NMR Structure of Putative N-Acetyl Transferase YhhK from E. coli Bound to Coenzyme A: Northeast Structural Genomics Consortium Target ET106 | | Descriptor: | COENZYME A, Uncharacterized protein yhhK | | Authors: | Cort, J.R, Yee, A, Montelione, G.T, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-30 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Putative N-Acetyl Transferase YhhK from E. coli Bound to Coenzyme A
To be Published
|
|
2KJ8
 
 | | NMR structure of fragment 87-196 from the putative phage integrase IntS of E. coli: Northeast Structural Genomics Consortium target ER652A, PSI-2 | | Descriptor: | Putative prophage CPS-53 integrase | | Authors: | Cort, J.R, Ramelot, T.A, Wang, D, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Swapna, G, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-25 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of fragment 87-196 from the putative phage integrase IntS of E. coli
To be Published
|
|
2HGK
 
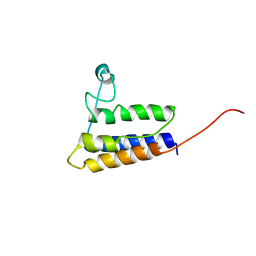 | | Solution NMR Structure of protein YqcC from E. coli: Northeast Structural Genomics Consortium target ER225 | | Descriptor: | Hypothetical protein yqcC | | Authors: | Cort, J.R, Wang, D, Shetty, K, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-27 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein YqcC from E.coli
To be Published
|
|
2HH8
 
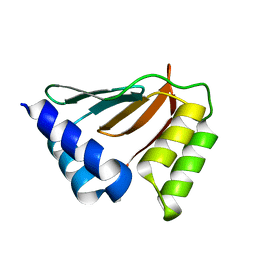 | | Solution NMR structure of the ydfO protein from Escherichia coli. Northeast Structural Genomics target ER251. | | Descriptor: | Hypothetical protein ydfO | | Authors: | Rossi, P, Cort, J.R, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-28 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the ydfO protein from Escherichia coli. Northeast Structural Genomics target ER251.
To be Published
|
|
