4LX9
 
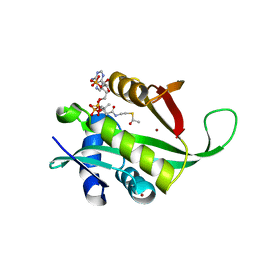 | |
4F2U
 
 | | Structure of the N254Y/H258Y double mutant of the Phosphatidylinositol-Specific Phospholipase C from S.aureus | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION | | Authors: | Cheng, J, Goldstein, R, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-05-08 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Competition between Anion Binding and Dimerization Modulates Staphylococcus aureus Phosphatidylinositol-specific Phospholipase C Enzymatic Activity.
J.Biol.Chem., 287, 2012
|
|
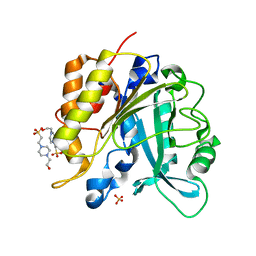
4F2T
 
 | | Modulation of S.aureus Phosphatidylinositol-Specific Phospholipase C Membrane Binding. | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | Cheng, J, Goldstein, R, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-05-08 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Competition between Anion Binding and Dimerization Modulates Staphylococcus aureus Phosphatidylinositol-specific Phospholipase C Enzymatic Activity.
J.Biol.Chem., 287, 2012
|
|
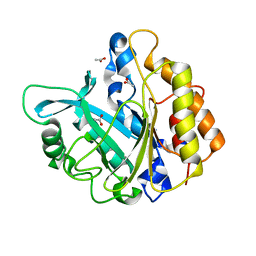
4F2B
 
 | | Modulation of S.Aureus Phosphatidylinositol-Specific Phospholipase C Membrane Binding | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase | | Authors: | Cheng, J, Goldstein, R, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-05-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Competition between Anion Binding and Dimerization Modulates Staphylococcus aureus Phosphatidylinositol-specific Phospholipase C Enzymatic Activity.
J.Biol.Chem., 287, 2012
|
|
2JFB
 
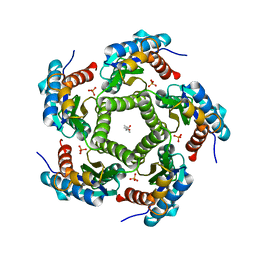 | | 3D Structure of Lumazine Synthase from Candida albicans | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, PHOSPHATE ION | | Authors: | Morgunova, E, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lumazine Synthase from Candida Albicans as an Anti- Fungal Target Enzyme: Structural and Biochemical Basis for Drug Design.
J.Biol.Chem., 282, 2007
|
|
3TFY
 
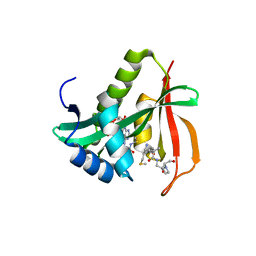 | |
4PO2
 
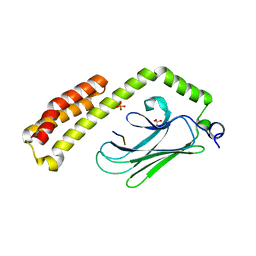 | | Crystal Structure of the Stress-Inducible Human Heat Shock Protein HSP70 Substrate-Binding Domain in Complex with Peptide Substrate | | Descriptor: | HSP70 substrate peptide, Heat shock 70 kDa protein 1A/1B, PHOSPHATE ION, ... | | Authors: | Zhang, P, Leu, J.I, Murphy, M.E, George, D.L, Marmorstein, R. | | Deposit date: | 2014-02-24 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the stress-inducible human heat shock protein 70 substrate-binding domain in complex with Peptide substrate.
Plos One, 9, 2014
|
|
4K2J
 
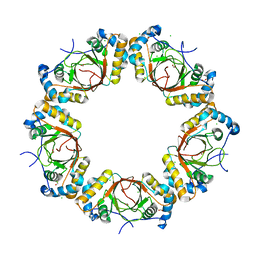 | |
1HU8
 
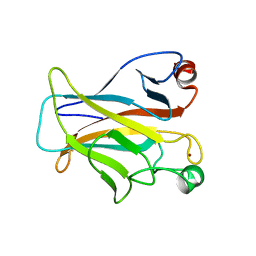 | | CRYSTAL STRUCTURE OF THE MOUSE P53 CORE DNA-BINDING DOMAIN AT 2.7A RESOLUTION | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Zhao, K, Chai, X, Johnston, K, Clements, A, Marmorstein, R. | | Deposit date: | 2001-01-04 | | Release date: | 2001-07-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the mouse p53 core DNA-binding domain at 2.7 A resolution.
J.Biol.Chem., 276, 2001
|
|
1HWT
 
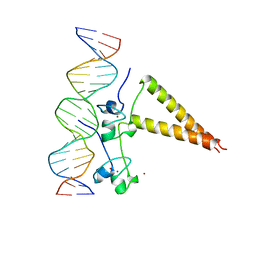 | | STRUCTURE OF A HAP1/DNA COMPLEX REVEALS DRAMATICALLY ASYMMETRIC DNA BINDING BY A HOMODIMERIC PROTEIN | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*TP*AP*TP*TP*AP*TP*CP*GP*CP*TP*AP*TP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*AP*TP*AP*GP*CP*GP*AP*TP*AP*AP*TP*AP*GP*CP*GP*C)-3'), PROTEIN (HEME ACTIVATOR PROTEIN), ... | | Authors: | King, D.A, Zhang, L, Guarente, L, Marmorstein, R. | | Deposit date: | 1998-09-17 | | Release date: | 1999-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a HAP1-DNA complex reveals dramatically asymmetric DNA binding by a homodimeric protein.
Nat.Struct.Biol., 6, 1999
|
|
4E26
 
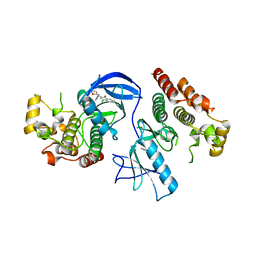 | | BRAF in complex with an organic inhibitor 7898734 | | Descriptor: | 5-chloro-7-[(R)-furan-2-yl(pyridin-2-ylamino)methyl]quinolin-8-ol, Serine/threonine-protein kinase B-raf | | Authors: | Qin, J, Xie, P, Ventocilla, C, Zhou, G, Vultur, A, Chen, Q, Herlyn, M, Winkler, J, Marmorstein, R. | | Deposit date: | 2012-03-07 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of a Novel Family of BRAF(V600E) Inhibitors.
J.Med.Chem., 55, 2012
|
|
4GS4
 
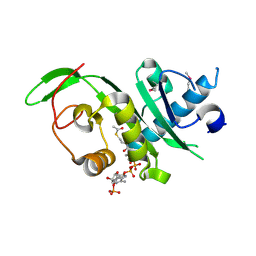 | | Structure of the alpha-tubulin acetyltransferase, alpha-TAT1 | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase | | Authors: | Friedmann, D.R, Fan, J, Marmorstein, R. | | Deposit date: | 2012-08-27 | | Release date: | 2012-10-17 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Structure of the alpha-tubulin acetyltransferase, alpha TAT1, and implications for tubulin-specific acetylation.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1HXH
 
 | | COMAMONAS TESTOSTERONI 3BETA/17BETA HYDROXYSTEROID DEHYDROGENASE | | Descriptor: | 3BETA/17BETA-HYDROXYSTEROID DEHYDROGENASE | | Authors: | Benach, J, Filling, C, Oppermann, U.C.T, Roversi, P, Bricogne, G, Berndt, K.D, Jornvall, H, Ladenstein, R. | | Deposit date: | 2001-01-15 | | Release date: | 2002-12-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structure of Bacterial 3beta/17beta-Hydroxysteroid Dehydrogenase at 1.2 A Resolution: A Model for
Multiple Steroid Recognition
Biochemistry, 41, 2002
|
|
1HQK
 
 | | CRYSTAL STRUCTURE ANALYSIS OF LUMAZINE SYNTHASE FROM AQUIFEX AEOLICUS | | Descriptor: | 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE | | Authors: | Zhang, X, Meining, W, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2000-12-18 | | Release date: | 2001-12-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile Aquifex aeolicus at 1.6 A resolution: determinants of thermostability revealed from structural comparisons.
J.Mol.Biol., 306, 2001
|
|
4EQC
 
 | | Crystal structure of PAK1 kinase domain in complex with FRAX597 inhibitor | | Descriptor: | 6-[2-chloro-4-(1,3-thiazol-5-yl)phenyl]-8-ethyl-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, CHLORIDE ION, Serine/threonine-protein kinase PAK 1 | | Authors: | Maksimoska, J, Marmorstein, R. | | Deposit date: | 2012-04-18 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | FRAX597, a Small Molecule Inhibitor of the p21-activated Kinases, Inhibits Tumorigenesis of Neurofibromatosis Type 2 (NF2)-associated Schwannomas.
J.Biol.Chem., 288, 2013
|
|
4PZT
 
 | | Crystal structure of p300 histone acetyltransferase domain in complex with an inhibitor, Acetonyl-Coenzyme A | | Descriptor: | DIMETHYL SULFOXIDE, Histone acetyltransferase p300, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-3-HYDROXY-2,2-DIMETHYL-4-OXO-4-{[3-OXO-3-({2-[(2-OXOPROPYL)THIO]ETHYL}AMINO)PROPYL]AMINO}BUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Maksimoska, J, Marmorstein, R. | | Deposit date: | 2014-03-31 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the p300 Histone Acetyltransferase Bound to Acetyl-Coenzyme A and Its Analogues.
Biochemistry, 53, 2014
|
|
1FY7
 
 | | CRYSTAL STRUCTURE OF YEAST ESA1 HISTONE ACETYLTRANSFERASE DOMAIN COMPLEXED WITH COENZYME A | | Descriptor: | COENZYME A, ESA1 HISTONE ACETYLTRANSFERASE, SODIUM ION | | Authors: | Yan, Y, Barlev, N.A, Haley, R.H, Berger, S.L, Marmorstein, R. | | Deposit date: | 2000-09-28 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases.
Mol.Cell, 6, 2000
|
|
4PZR
 
 | |
4PZS
 
 | |
1MJB
 
 | | Crystal structure of yeast Esa1 histone acetyltransferase E338Q mutant complexed with acetyl coenzyme A | | Descriptor: | ACETYL COENZYME *A, Esa1 protein | | Authors: | Yan, Y, Harper, S, Speicher, D, Marmorstein, R. | | Deposit date: | 2002-08-27 | | Release date: | 2002-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate.
Nat.Struct.Biol., 9, 2002
|
|
1N1Q
 
 | | Crystal structure of a Dps protein from Bacillus brevis | | Descriptor: | DPS Protein, MU-OXO-DIIRON | | Authors: | Ren, B, Tibbelin, G, Kajino, T, Asami, O, Ladenstein, R. | | Deposit date: | 2002-10-19 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Multi-layered Structure of Dps with a Novel Di-nuclear Ferroxidase Center
J.Mol.Biol., 329, 2003
|
|
1NFJ
 
 | | Structure of a Sir2 substrate, alba, reveals a mechanism for deactylation-induced enhancement of DNA-binding | | Descriptor: | conserved hypothetical protein AF1956 | | Authors: | Zhao, K, Chai, X, Marmorstein, R. | | Deposit date: | 2002-12-15 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Sir2 substrate, alba, reveals a mechanism for deacetylation-induced enhancement of DNA-binding
J.Biol.Chem., 278, 2003
|
|
3Q4C
 
 | | Crystal Structure of Wild Type BRAF kinase domain in complex with organometallic inhibitor CNS292 | | Descriptor: | Serine/threonine-protein kinase B-raf, [(1,2,3,4,5,6-eta)-(1S,2R,3R,4R,5S,6S)-1-carboxycyclohexane-1,2,3,4,5,6-hexayl](chloro)(3-methyl-5,7-dioxo-6,7-dihydro-5H-pyrido[2,3-a]pyrrolo[3,4-c]carbazol-12-ide-kappa~2~N~1~,N~12~)ruthenium(1+) | | Authors: | Xie, P, Streu, C, Qin, J, Pregman, H, Pagano, N, Meggers, E, Marmorstein, R. | | Deposit date: | 2010-12-23 | | Release date: | 2011-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of BRAF in complex with an organoruthenium inhibitor reveals a mechanism for inhibition of an active form of BRAF kinase.
Biochemistry, 48, 2009
|
|
1NQV
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 5-nitroso-6-ribityl-amino-2,4(1H,3H)pyrimidinedione | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
3QM0
 
 | | Crystal structure of RTT109-AC-CoA complex | | Descriptor: | ACETYL COENZYME *A, Histone acetyltransferase RTT109, MERCURY (II) ION | | Authors: | Tang, Y, Marmorstein, R. | | Deposit date: | 2011-02-03 | | Release date: | 2011-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP.
Nat.Struct.Mol.Biol., 15, 2008
|
|
