5LNV
 
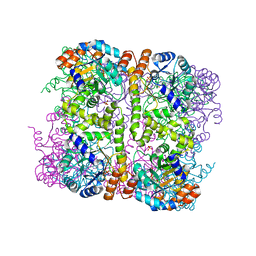 | | Crystal structure of Arabidopsis thaliana Pdx1-I320 complex from multiple crystals | | Descriptor: | (4~{S})-4-azanyl-5-oxidanyl-pent-1-en-3-one, PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
5LTQ
 
 | |
5LNT
 
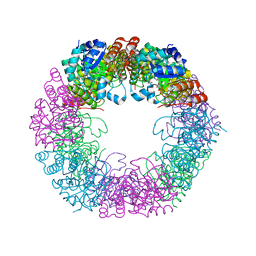 | | Crystal structure of Arabidopsis thaliana Pdx1K166R-preI320 complex | | Descriptor: | PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.1, [(~{E},4~{S})-4-azanyl-3-oxidanylidene-pent-1-enyl] dihydrogen phosphate | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
5LNW
 
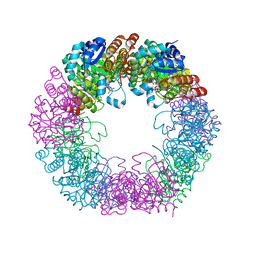 | | Crystal structure of Arabidopsis thaliana Pdx1-I320-G3P complex | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, GLYCEROL, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
5AJG
 
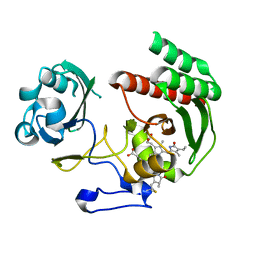 | | Structure of Infrared Fluorescent Protein IFP1.4 AT 1.11 Angstrom resolution | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, BACTERIOPHYTOCHROME | | Authors: | Lafaye, C, Shu, X, Royant, A. | | Deposit date: | 2015-02-24 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structural Determinants of Improved Fluorescence in a Family of Bacteriophytochrome-Based Infrared Fluorescent Proteins: Insights from Continuum Electrostatic Calculations and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
5LTR
 
 | |
5M7A
 
 | |
5M79
 
 | |
1TWQ
 
 | | Crystal structure of the C-terminal PGN-binding domain of human PGRP-Ialpha in complex with PGN analog muramyl tripeptide | | Descriptor: | N-acetyl-beta-muramic acid, NICKEL (II) ION, muramyl tripeptide, ... | | Authors: | Guan, R, Roychowdury, A, Boons, G.-A, Mariuzza, R.A. | | Deposit date: | 2004-07-01 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for peptidoglycan binding by peptidoglycan recognition proteins
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1QHJ
 
 | | X-RAY STRUCTURE OF BACTERIORHODOPSIN GROWN IN LIPIDIC CUBIC PHASES | | Descriptor: | 1,2-[DI-2,6,10,14-TETRAMETHYL-HEXADECAN-16-OXY]-PROPANE, PROTEIN (BACTERIORHODOPSIN), RETINAL | | Authors: | Belrhali, H, Nollert, P, Royant, A, Menzel, C, Rosenbusch, J.P, Landau, E.M, Pebay-Peyroula, E. | | Deposit date: | 1999-05-04 | | Release date: | 1999-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein, lipid and water organization in bacteriorhodopsin crystals: a molecular view of the purple membrane at 1.9 A resolution.
Structure Fold.Des., 7, 1999
|
|
6GWZ
 
 | |
6GX0
 
 | |
6GPU
 
 | | Crystal structure of miniSOG at 1.17A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Lafaye, C, Signor, L, Aumonier, S, Shu, X, Gotthard, G, Royant, A. | | Deposit date: | 2018-06-07 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Tailing miniSOG: structural bases of the complex photophysics of a flavin-binding singlet oxygen photosensitizing protein.
Sci Rep, 9, 2019
|
|
6GX1
 
 | |
6GX2
 
 | |
6GPV
 
 | | Crystal structure of blue-light irradiated miniSOG | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, LUMICHROME, ... | | Authors: | Lafaye, C, Signor, L, Aumonier, S, Shu, X, Gotthard, G, Royant, A. | | Deposit date: | 2018-06-07 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tailing miniSOG: structural bases of the complex photophysics of a flavin-binding singlet oxygen photosensitizing protein.
Sci Rep, 9, 2019
|
|
6GWY
 
 | |
2B6M
 
 | |
2B3S
 
 | |
2XBS
 
 | | Raman crystallography of Hen White Egg Lysozyme - High X-ray dose (16 MGy) | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Carpentier, P, Royant, A, Weik, M, Bourgeois, D. | | Deposit date: | 2010-04-14 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Raman Assisted Crystallography Reveals a Mechanism of X-Ray Induced Reversible Disulfide Radical Formation
Structure, 18, 2010
|
|
2XBR
 
 | | Raman crystallography of Hen White Egg Lysozyme - Low X-ray dose (0.2 MGy) | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Carpentier, P, Royant, A, Weik, M, Bourgeois, D. | | Deposit date: | 2010-04-14 | | Release date: | 2010-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Raman Assisted Crystallography Reveals a Mechanism of X-Ray Induced Reversible Disulfide Radical Formation
Structure, 18, 2010
|
|
2YE1
 
 | | X-ray structure of the cyan fluorescent proteinmTurquoise-GL (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN, MAGNESIUM ION | | Authors: | von Stetten, D, Noirclerc-Savoye, M, Goedhart, J, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-03-25 | | Release date: | 2012-04-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Characterization of the Cyan Fluorescent Protein Mturquoise-Gl
To be Published
|
|
2ZJZ
 
 | | Structure of the K349P mutant of Gi alpha 1 subunit bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit | | Authors: | Morikawa, T, Muroya, A, Sugio, S, Wakamatsu, K, Kohno, T. | | Deposit date: | 2008-03-11 | | Release date: | 2009-03-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | How GPCRs activate G proteins: Structural changes from C-terminal tail to GDP binding pocket
To be Published
|
|
2ZJY
 
 | | Structure of the K349P mutant of Gi alpha 1 subunit bound to ALF4 and GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Morikawa, T, Muroya, A, Sugio, S, Wakamatsu, K, Kohno, T. | | Deposit date: | 2008-03-11 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | How GPCRs activate G proteins: Structural changes form C-terminal tail to GDP binding pocket
To be Published
|
|
3B7S
 
 | | [E296Q]LTA4H in complex with RSR substrate | | Descriptor: | ACETIC ACID, GLYCEROL, Leukotriene A-4 hydrolase, ... | | Authors: | Tholander, F, Haeggstrom, J, Thunnissen, M, Muroya, A, Roques, B.-P, Fournie-Zaluski, M.-C. | | Deposit date: | 2007-10-31 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.465 Å) | | Cite: | Structure-based dissection of the active site chemistry of leukotriene a4 hydrolase: implications for m1 aminopeptidases and inhibitor design.
Chem.Biol., 15, 2008
|
|
