2GVQ
 
 | | Anthranilate phosphoribosyl-transferase (TRPD) from S. solfataricus in complex with anthranilate | | Descriptor: | 2-AMINOBENZOIC ACID, Anthranilate phosphoribosyltransferase | | Authors: | Marino, M, Deuss, M, Sterner, R, Mayans, O. | | Deposit date: | 2006-05-03 | | Release date: | 2006-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from Sulfolobus solfataricus.
J.Biol.Chem., 281, 2006
|
|
2I5P
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase isoform 1 from K. marxianus | | Descriptor: | BETA-MERCAPTOETHANOL, Glyceraldehyde-3-phosphate dehydrogenase 1, alpha-D-glucopyranose | | Authors: | Ferreira-da-Silva, F, Pereira, P.J.B, Gales, L, Moradas-Ferreira, P, Damas, A.M. | | Deposit date: | 2006-08-25 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal and Solution Structures of Glyceraldehyde-3-phosphate Dehydrogenase Reveal Different Quaternary Structures.
J.Biol.Chem., 281, 2006
|
|
1WT6
 
 | | Coiled-Coil domain of DMPK | | Descriptor: | Myotonin-protein kinase | | Authors: | Garcia, P, Marino, M, Mayans, O. | | Deposit date: | 2004-11-16 | | Release date: | 2005-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insights into the self-assembly mechanism of dystrophia myotonica kinase
Faseb J., 20, 2006
|
|
1YRP
 
 | |
1YS7
 
 | | Crystal structure of the response regulator protein prrA complexed with Mg2+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Nowak, E, Panjikar, S, Tucker, P, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2005-02-07 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The structural basis of signal transduction for the response regulator PrrA from Mycobacterium tuberculosis.
J.Biol.Chem., 281, 2006
|
|
1ZYK
 
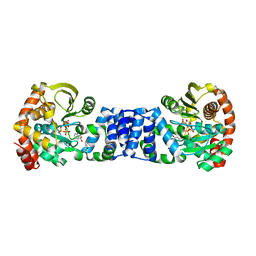 | | Anthranilate Phosphoribosyltransferase in complex with PRPP, anthranilate and magnesium | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-AMINOBENZOIC ACID, Anthranilate phosphoribosyltransferase, ... | | Authors: | Marino, M, Deuss, M, Sterner, R, Mayans, O. | | Deposit date: | 2005-06-10 | | Release date: | 2006-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from Sulfolobus solfataricus.
J.Biol.Chem., 281, 2006
|
|
1ZXY
 
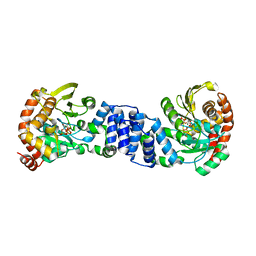 | | Anthranilate Phosphoribosyltransferase from Sulfolobus solfataricus in complex with PRPP and Magnesium | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Anthranilate phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Marino, M, Deuss, M, Sterner, R, Mayans, O. | | Deposit date: | 2005-06-09 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from Sulfolobus solfataricus.
J.Biol.Chem., 281, 2006
|
|
1ZAN
 
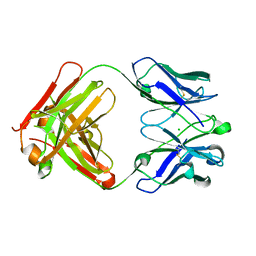 | | Crystal structure of anti-NGF AD11 Fab | | Descriptor: | CHLORIDE ION, Fab AD11 Heavy Chain, Fab AD11 Light Chain | | Authors: | Covaceuszach, S, Cattaneo, A, Cassetta, A, Lamba, D. | | Deposit date: | 2005-04-06 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dissecting NGF interactions with TrkA and p75 receptors by structural and functional studies of an anti-NGF neutralizing antibody.
J.Mol.Biol., 381, 2008
|
|
2A2A
 
 | |
2XZS
 
 | | Death associated protein kinase 1 residues 1-312 | | Descriptor: | DEATH ASSOCIATED KINASE 1, MAGNESIUM ION | | Authors: | Yumerefendi, H, Mas, P.J, Dordevic, N, McCarthy, A.A, Hart, D.J. | | Deposit date: | 2010-11-29 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Death-Associated Protein Kinase Activity is Regulated by Coupled Calcium/Calmodulin Binding to Two Distinct Sites.
Structure, 24, 2016
|
|
2Y25
 
 | |
3CA3
 
 | | Crystal structure of Sambucus Nigra Agglutinin II (SNA-II)-tetragonal crystal form- complexed to N-Acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maveyraud, L, Mourey, L. | | Deposit date: | 2008-02-19 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
3CA6
 
 | | Sambucus nigra agglutinin II (SNA-II)- tetragonal crystal form- complexed to Tn antigen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maveyraud, L, Mourey, L. | | Deposit date: | 2008-02-19 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
8AFO
 
 | | Structure of fibronectin 2 and 3 of L1CAM at 2.0 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Neural cell adhesion molecule L1 | | Authors: | Guedez, G, Loew, C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | X-ray structure and function of fibronectin domains two and three of the neural cell adhesion molecule L1.
Faseb J., 37, 2023
|
|
8BGP
 
 | | N,N-diacetylchitobiose deacetylase from Pyrococcus chitonophagus anomalous data | | Descriptor: | Diacetylchitobiose deacetylase, ZINC ION | | Authors: | Rypniewski, W, Biniek-Antosiak, K, Bejger, M. | | Deposit date: | 2022-10-28 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural, Thermodynamic and Enzymatic Characterization of N , N -Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus.
Int J Mol Sci, 23, 2022
|
|
8BGO
 
 | | N,N-diacetylchitobiose deacetylase from Pyrococcus chitonophagus with substrate N,N-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Diacetylchitobiose deacetylase, ZINC ION | | Authors: | Rypniewski, W, Bejger, M, Biniek-Antosiak, K. | | Deposit date: | 2022-10-28 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural, Thermodynamic and Enzymatic Characterization of N , N -Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus.
Int J Mol Sci, 23, 2022
|
|
8BGN
 
 | | N,N-diacetylchitobiose deacetylase from Pyrococcus chitonophagus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Diacetylchitobiose deacetylase, ... | | Authors: | Rypniewski, W, Bejger, M, Biniek-Antosiak, K. | | Deposit date: | 2022-10-28 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural, Thermodynamic and Enzymatic Characterization of N , N -Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus.
Int J Mol Sci, 23, 2022
|
|
8AFP
 
 | | Structure of fibronectin 2 and 3 of L1CAM at 3.0 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neural cell adhesion molecule L1 | | Authors: | Guedez, G, Loew, C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure and function of fibronectin domains two and three of the neural cell adhesion molecule L1.
Faseb J., 37, 2023
|
|
8A17
 
 | | Human PTPRM domains FN3-4, in spacegroup P3221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase mu, ... | | Authors: | Shamin, M, Graham, S.C, Sharpe, H.J, Deane, J.E. | | Deposit date: | 2022-05-31 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Determinants of receptor tyrosine phosphatase homophilic adhesion: Structural comparison of PTPRK and PTPRM extracellular domains.
J.Biol.Chem., 299, 2023
|
|
8A1F
 
 | | Human PTPRK N-terminal domains MAM-Ig-FN1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hay, I.M, Graham, S.C, Sharpe, H.J, Deane, J.E. | | Deposit date: | 2022-06-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Determinants of receptor tyrosine phosphatase homophilic adhesion: Structural comparison of PTPRK and PTPRM extracellular domains.
J.Biol.Chem., 299, 2023
|
|
8A16
 
 | | Human PTPRM domains FN3-4, in spacegroup P212121 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase mu, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Caroe, E, Graham, S.C, Sharpe, H.J, Deane, J.E. | | Deposit date: | 2022-05-31 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Determinants of receptor tyrosine phosphatase homophilic adhesion: Structural comparison of PTPRK and PTPRM extracellular domains.
J.Biol.Chem., 299, 2023
|
|
4C9T
 
 | | BACTERIAL CHALCONE ISOMERASE IN open CONFORMATION FROM EUBACTERIUM RAMULUS AT 2.0 A RESOLUTION, SelenoMet derivative | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2013-10-03 | | Release date: | 2014-10-22 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4C9S
 
 | | BACTERIAL CHALCONE ISOMERASE IN open CONFORMATION FROM EUBACTERIUM RAMULUS AT 1.8 A RESOLUTION | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2013-10-03 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4D1E
 
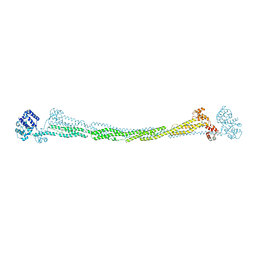 | | THE CRYSTAL STRUCTURE OF HUMAN MUSCLE ALPHA-ACTININ-2 | | Descriptor: | ALPHA-ACTININ-2 | | Authors: | Pinotsis, N, Salmazo, A, Sjoeblom, B, Gkougkoulia, E, Djinovic-Carugo, K. | | Deposit date: | 2014-05-01 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Structure and Regulation of Human Muscle Alpha-Actinin
Cell(Cambridge,Mass.), 159, 2014
|
|
4D06
 
 | | Bacterial chalcone isomerase complexed with naringenin | | Descriptor: | (2E)-3-(4-hydroxyphenyl)-1-(2,4,6-trihydroxyphenyl)prop-2-en-1-one, CHALCONE ISOMERASE, CHLORIDE ION, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-04-24 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
