4CE4
 
 | | 39S large subunit of the porcine mitochondrial ribosome | | Descriptor: | 16S Ribosomal RNA, ICT1, MRPL13, ... | | Authors: | Greber, B.J, Boehringer, D, Leitner, A, Bieri, P, Voigts-Hoffmann, F, Erzberger, J.P, Leibundgut, M, Aebersold, R, Ban, N. | | Deposit date: | 2013-11-08 | | Release date: | 2013-12-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Architecture of the Large Subunit of the Mammalian Mitochondrial Ribosome.
Nature, 505, 2014
|
|
4BTS
 
 | | THE CRYSTAL STRUCTURE OF THE EUKARYOTIC 40S RIBOSOMAL SUBUNIT IN COMPLEX WITH EIF1 AND EIF1A | | Descriptor: | 18S ribosomal RNA, 40S RIBOSOMAL PROTEIN RACK1, 40S RIBOSOMAL PROTEIN RPS10E, ... | | Authors: | Weisser, M, Voigts-Hoffmann, F, Rabl, J, Leibundgut, M, Ban, N. | | Deposit date: | 2013-06-19 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.703 Å) | | Cite: | The crystal structure of the eukaryotic 40S ribosomal subunit in complex with eIF1 and eIF1A.
Nat. Struct. Mol. Biol., 20, 2013
|
|
4BSZ
 
 | |
2KPD
 
 | | Structure determination of the top-loop of the conserved 3 terminal secondary structure in the genome of YFV-mutant | | Descriptor: | RNA (5'-R(*UP*GP*AP*GP*CP*UP*CP*AP*GP*UP*UP*UP*GP*CP*UP*CP*A)-3') | | Authors: | Lescrinier, E, Dyubankova, N, Nauwelaerts, K, Jones, R, Herdewijn, P. | | Deposit date: | 2009-10-12 | | Release date: | 2010-06-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of the Top-Loop of the Conserved 3'-Terminal Secondary Structure in the Genome of Flaviviruses.
Chembiochem, 11, 2010
|
|
6ZMX
 
 | | Crystal structure of hemoglobin from turkey (Meleagiris gallopova) crystallized in orthorhombic form at 1.4 Angstrom resolution | | Descriptor: | Hemoglobin beta chain, Hemoglobin subunit alpha-A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pandian, R, Shobana, N, Sundaresan, S.S, Sayed, Y, Ponnuswamy, M.N. | | Deposit date: | 2020-07-04 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.389 Å) | | Cite: | Structural studies of hemoglobin from two flightless birds, ostrich and turkey: insights into their differing oxygen-binding properties.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZMY
 
 | | Crystal structure of hemoglobin from turkey (Meleagiris gallopova) crystallized in monoclinic form at 1.66 Angstrom resolution | | Descriptor: | Hemoglobin beta chain, Hemoglobin subunit alpha-A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pandian, R, Shobana, N, Sundaresan, S.S, Thangaraj, V, Sayed, Y, Ponnuswamy, M.N. | | Deposit date: | 2020-07-04 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Structural studies of hemoglobin from two flightless birds, ostrich and turkey: insights into their differing oxygen-binding properties.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZZG
 
 | | MB_CRS6-1 bound to CrSAS-6_N | | Descriptor: | ACETATE ION, Centriole protein, MB_CRS6-15 | | Authors: | Hatzopoulos, G.N, Kukenshoner, T, Banterle, N, Favez, T, Fluckiger, I, Hantschel, O, Gonczy, P. | | Deposit date: | 2020-08-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Tuning SAS-6 architecture with monobodies impairs distinct steps of centriole assembly.
Nat Commun, 12, 2021
|
|
6ZZ8
 
 | | MB_CRS6-15 bound to CrSAS-6_6HR | | Descriptor: | Centriole protein, Protein B | | Authors: | Hatzopoulos, G.N, Kukenshoner, T, Banterle, N, Favez, T, Fluckiger, I, Hantschel, O, Gonczy, P. | | Deposit date: | 2020-08-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | Tuning SAS-6 architecture with monobodies impairs distinct steps of centriole assembly.
Nat Commun, 12, 2021
|
|
6ZZD
 
 | | MB_CRS6-13 bound to CrSAS-6_N | | Descriptor: | Centriole protein, MB_CRS6-13 | | Authors: | Hatzopoulos, G.N, Kukenshoner, T, Banterle, N, Favez, T, Fluckiger, I, Hantschel, O, Gonczy, P. | | Deposit date: | 2020-08-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Tuning SAS-6 architecture with monobodies impairs distinct steps of centriole assembly.
Nat Commun, 12, 2021
|
|
6ZZC
 
 | | MB_CRS6-1 bound to CrSAS-6_6HR | | Descriptor: | Centriole protein, DODECAETHYLENE GLYCOL, MB_CrS6-1 | | Authors: | Hatzopoulos, G.N, Kukenshoner, T, Banterle, N, Favez, T, Fluckiger, I, Hantschel, O, Gonczy, P. | | Deposit date: | 2020-08-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Tuning SAS-6 architecture with monobodies impairs distinct steps of centriole assembly.
Nat Commun, 12, 2021
|
|
5LMZ
 
 | | Fluorinase from Streptomyces sp. MA37 | | Descriptor: | 1-DEAZA-ADENOSINE, CHLORIDE ION, Fluorinase, ... | | Authors: | Mann, G, O'Hagan, D, Naismith, J.H, Bandaranayaka, N. | | Deposit date: | 2016-08-02 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of fluorinases from Streptomyces sp MA37, Norcardia brasiliensis, and Actinoplanes sp N902-109 by genome mining.
Chembiochem, 15, 2014
|
|
5A9Q
 
 | | Human nuclear pore complex | | Descriptor: | NUCLEAR PORE COMPLEX PROTEIN NUP107, NUCLEAR PORE COMPLEX PROTEIN NUP133, NUCLEAR PORE COMPLEX PROTEIN NUP155, ... | | Authors: | von Appen, A, Kosinski, J, Sparks, L, Ori, A, DiGuilio, A, Vollmer, B, Mackmull, M, Banterle, N, Parca, L, Kastritis, P, Buczak, K, Mosalaganti, S, Hagen, W, Andres-Pons, A, Lemke, E.A, Bork, P, Antonin, W, Glavy, J.S, Bui, K.H, Beck, M. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | In Situ Structural Analysis of the Human Nuclear Pore Complex
Nature, 526, 2015
|
|
5C4L
 
 | | Conformational alternate of sisomicin in complex with APH(2")-IVa | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, (2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-ol, APH(2'')-Id | | Authors: | Kaplan, E, Guichou, J.F, Berrou, K, Chaloin, L, Leban, N, Lallemand, P, Barman, T, Serpersu, E.H, Lionne, C. | | Deposit date: | 2015-06-18 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Aminoglycoside binding and catalysis specificity of aminoglycoside 2-phosphotransferase IVa: A thermodynamic, structural and kinetic study.
Biochim.Biophys.Acta, 1860, 2016
|
|
5C4K
 
 | | APH(2")-IVa in complex with GET (G418) at room temperature | | Descriptor: | APH(2'')-Id, GENETICIN | | Authors: | Kaplan, E, Guichou, J.F, Berrou, K, Chaloin, L, Leban, N, Lallemand, P, Barman, T, Serpersu, E.H, Lionne, C. | | Deposit date: | 2015-06-18 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Aminoglycoside binding and catalysis specificity of aminoglycoside 2-phosphotransferase IVa: A thermodynamic, structural and kinetic study.
Biochim.Biophys.Acta, 1860, 2016
|
|
2KPC
 
 | | Structure determination of the top-loop of the conserved 3 terminal secondary structure in the genome of YFV | | Descriptor: | RNA (5'-R(*UP*GP*AP*GP*CP*AP*CP*AP*GP*UP*UP*UP*GP*CP*UP*CP*A)-3') | | Authors: | Lescrinier, E, Dyubankova, N, Nauwelaerts, K, Jones, R, Herdewijn, P. | | Deposit date: | 2009-10-12 | | Release date: | 2010-06-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of the Top-Loop of the Conserved 3'-Terminal Secondary Structure in the Genome of Flaviviruses.
Chembiochem, 11, 2010
|
|
4J2H
 
 | | Crystal structure of a putative short-chain alcohol dehydrogenase from Sinorhizobium meliloti 1021 (Target NYSGRC-011708) | | Descriptor: | 1,2-ETHANEDIOL, PENTAETHYLENE GLYCOL, SODIUM ION, ... | | Authors: | Sampathkumar, P, Gizzi, A, Ahmed, M, Banu, N, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Fiser, A, Glenn, A.S, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Love, J.D, Stead, M, Seidel, R, Toro, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a putative short-chain alcohol dehydrogenase from Sinorhizobium meliloti 1021 (Target NYSGRC-011708)
to be published
|
|
4N57
 
 | | Crystal structure of aminoglycoside phosphotransferase APH(2'')-IVa ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, APH(2'')-Id, MAGNESIUM ION | | Authors: | Kaplan, E, Leban, N, Chaloin, L, Guichou, J.-F, Lionne, C. | | Deposit date: | 2013-10-09 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of aminoglycoside phosphotransferase APH(2'')-IVa ADP complex
To be Published
|
|
3KIO
 
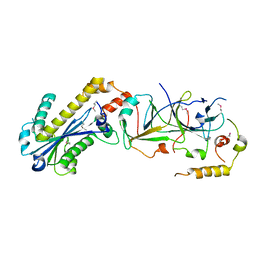 | | mouse RNase H2 complex | | Descriptor: | Ribonuclease H2 subunit A, Ribonuclease H2 subunit B, Ribonuclease H2 subunit C | | Authors: | Shaban, N, Harvey, S, Perrino, F.W, Hollis, T. | | Deposit date: | 2009-11-02 | | Release date: | 2009-11-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of the mammalian RNase H2 complex provides insight into RNA:DNA hybrid processing to prevent immune dysfunction.
J.Biol.Chem., 285, 2010
|
|
8BUU
 
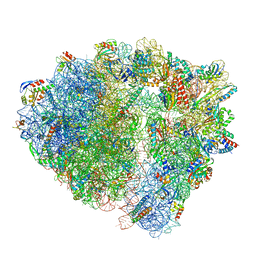 | | ARE-ABCF VmlR2 bound to a 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2022-11-30 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Genome-encoded ABCF factors implicated in intrinsic antibiotic resistance in Gram-positive bacteria: VmlR2, Ard1 and CplR.
Nucleic Acids Res., 51, 2023
|
|
4V4W
 
 | |
7OYF
 
 | |
7OYH
 
 | |
7OY3
 
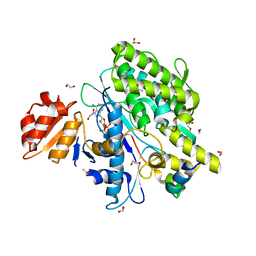 | | Crystal structure of depupylase Dop in complex with phosphorylated Pup and ADP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Cui, H. | | Deposit date: | 2021-06-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structures of prokaryotic ubiquitin-like protein Pup in complex with depupylase Dop reveal the mechanism of catalytic phosphate formation.
Nat Commun, 12, 2021
|
|
7OXY
 
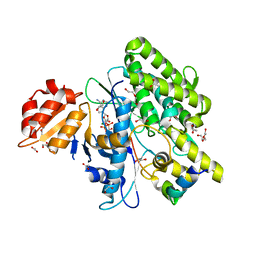 | | Crystal structure of depupylase Dop in complex with Pup and AMP-PCP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cui, H. | | Deposit date: | 2021-06-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of prokaryotic ubiquitin-like protein Pup in complex with depupylase Dop reveal the mechanism of catalytic phosphate formation.
Nat Commun, 12, 2021
|
|
7OXV
 
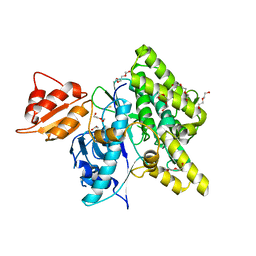 | | Crystal structure of depupylase Dop in the Dop-loop-inserted state | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Depupylase, ... | | Authors: | Cui, H. | | Deposit date: | 2021-06-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.394 Å) | | Cite: | Structures of prokaryotic ubiquitin-like protein Pup in complex with depupylase Dop reveal the mechanism of catalytic phosphate formation.
Nat Commun, 12, 2021
|
|
