3GKX
 
 | |
3GKU
 
 | | Crystal structure of a probable RNA-binding protein from Clostridium symbiosum ATCC 14940 | | Descriptor: | Probable RNA-binding protein | | Authors: | Tan, K, Keigher, L, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-11 | | Release date: | 2009-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of a probable RNA-binding protein from Clostridium symbiosum ATCC 14940
To be Published
|
|
2QRR
 
 | | Crystal structure of the soluble domain of the ABC transporter, ATP-binding protein from Vibrio parahaemolyticus | | Descriptor: | CHLORIDE ION, Methionine import ATP-binding protein metN | | Authors: | Kim, Y, Zhou, M, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-28 | | Release date: | 2007-08-14 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Soluble Domain of the ABC Transporter, ATP-binding Protein from Vibrio parahaemolyticus.
To be Published
|
|
2R2Z
 
 | | The crystal structure of a hemolysin domain from Enterococcus faecalis V583 | | Descriptor: | Hemolysin, ZINC ION | | Authors: | Zhang, R, Tan, K, Zhou, M, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-28 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of a hemolysin domain from Enterococcus faecalis V583.
To be Published
|
|
2R41
 
 | |
3GVE
 
 | | Crystal structure of calcineurin-like phosphoesterase YfkN from Bacillus subtilis | | Descriptor: | CITRIC ACID, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Kim, Y, Marshall, N, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-30 | | Release date: | 2009-04-28 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Calcineurin-like Phosphoesterase from Bacillus subtilis
To be Published
|
|
2R2C
 
 | |
3H2Z
 
 | |
2QM3
 
 | | Crystal structure of a predicted methyltransferase from Pyrococcus furiosus | | Descriptor: | ACETIC ACID, CALCIUM ION, Predicted methyltransferase | | Authors: | Cuff, M.E, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-13 | | Release date: | 2007-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of a predicted methyltransferase from Pyrococcus furiosus.
TO BE PUBLISHED
|
|
2QMM
 
 | |
2QH9
 
 | |
2QHK
 
 | |
2QH1
 
 | | Structure of TA289, a CBS-rubredoxin-like protein, in its Fe+2-bound state | | Descriptor: | FE (II) ION, Hypothetical protein Ta0289 | | Authors: | Singer, A.U, Proudfoot, M, Brown, G, Xu, L, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-06-29 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural characterization of a novel family of cystathionine beta-synthase domain proteins fused to a Zn ribbon-like domain.
J.Mol.Biol., 375, 2008
|
|
2QKO
 
 | | Crystal structure of transcriptional regulator RHA06399 from Rhodococcus sp. RHA1 | | Descriptor: | Possible transcriptional regulator, TetR family protein | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-11 | | Release date: | 2007-07-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of tetR family protein.
To be Published
|
|
3GL5
 
 | | Crystal structure of probable DsbA oxidoreductase SCO1869 from Streptomyces coelicolor | | Descriptor: | ACETATE ION, Putative DsbA oxidoreductase SCO1869, SODIUM ION | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-11 | | Release date: | 2009-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of probable DsbA oxidoreductase SCO1869 from Streptomyces coelicolor
To be Published
|
|
2QMA
 
 | | Crystal structure of glutamate decarboxylase domain of diaminobutyrate-pyruvate transaminase and L-2,4-diaminobutyrate decarboxylase from Vibrio parahaemolyticus | | Descriptor: | 1,2-ETHANEDIOL, Diaminobutyrate-pyruvate transaminase and L-2,4-diaminobutyrate decarboxylase | | Authors: | Osipiuk, J, Sather, A, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-14 | | Release date: | 2007-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray crystal structure of glutamate decarboxylase domain of diaminobutyrate-pyruvate transaminase and L-2,4-diaminobutyrate decarboxylase from Vibrio parahaemolyticus.
To be Published
|
|
3GMI
 
 | |
2QMO
 
 | | Crystal structure of dethiobiotin synthetase (bioD) from Helicobacter pylori | | Descriptor: | CHLORIDE ION, Dethiobiotin synthetase | | Authors: | Chruszcz, M, Xu, X, Cuff, M, Cymborowski, M, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-16 | | Release date: | 2007-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
2QQY
 
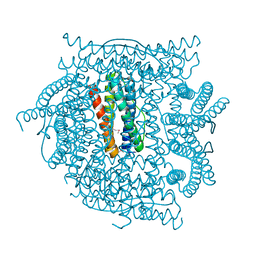 | | Crystal structure of ferritin like, diiron-carboxylate proteins from Bacillus anthracis str. Ames | | Descriptor: | Sigma B operon | | Authors: | Kim, Y, Joachimiak, G, Wu, R, Patterson, S, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-14 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Ferritin like, Diiron-carboxylate Proteins from Bacillus anthracis str. Ames.
To be Published
|
|
2QSA
 
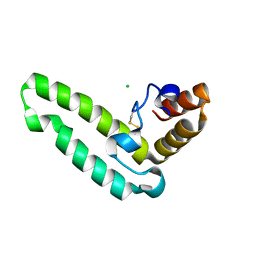 | | Crystal structure of J-domain of DnaJ homolog dnj-2 precursor from C.elegans. | | Descriptor: | CHLORIDE ION, DnaJ homolog dnj-2 | | Authors: | Osipiuk, J, Mulligan, R, Gu, M, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray crystal structure of J-domain of DnaJ homolog dnj-2 precursor from C.elegans.
To be Published
|
|
3H9P
 
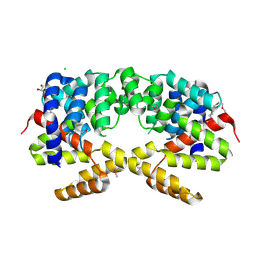 | | Crystal structure of putative triphosphoribosyl-dephospho-coA synthase from Archaeoglobus fulgidus | | Descriptor: | CHLORIDE ION, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Chang, C, Wu, R, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-30 | | Release date: | 2009-05-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of putative triphosphoribosyl-dephospho-coA synthase from Archaeoglobus fulgidus
To be Published
|
|
3HB7
 
 | | The Crystal Structure of an Isochorismatase-like Hydrolase from Alkaliphilus metalliredigens to 2.3A | | Descriptor: | AMMONIUM ION, Isochorismatase hydrolase, SODIUM ION | | Authors: | Stein, A.J, Xu, X, Cui, H, Ng, J, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-04 | | Release date: | 2009-07-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of an Isochorismatase-like Hydrolase from Alkaliphilus metalliredigens to 2.3A
To be Published
|
|
2QYB
 
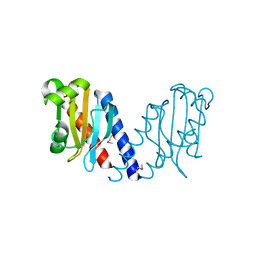 | |
3EUP
 
 | | The crystal structure of the transcriptional regulator, TetR family from Cytophaga hutchinsonii | | Descriptor: | SULFATE ION, Transcriptional regulator, TetR family | | Authors: | Zhang, R, Bigelow, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-10 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the transcriptional regulator, TetR family from Cytophaga hutchinsonii
To be Published
|
|
2QLT
 
 | | Crystal structure of an isoform of DL-glycerol-3-phosphatase, Rhr2p, from Saccharomyces cerevisiae | | Descriptor: | (DL)-glycerol-3-phosphatase 1, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-13 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of an isoform of DL-glycerol-3-phosphatase, Rhr2p from Saccharomyces cerevisiae.
To be Published
|
|
