1IE3
 
 | | CRYSTAL STRUCTURE OF R153C E. COLI MALATE DEHYDROGENASE | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRUVIC ACID | | Authors: | Bell, J.K, Yennawar, H.P, Wright, S.K, Thompson, J.R, Viola, R.E, Banaszak, L.J. | | Deposit date: | 2001-04-05 | | Release date: | 2001-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analyses of a Malate Dehydrogenase with a Variable Active Site
J.Biol.Chem., 276, 2001
|
|
5JN2
 
 | | Crystal structure of TgCDPK1 bound to NVPACU106 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calmodulin-domain protein kinase 1, ... | | Authors: | El Bakkouri, M, Walker, J.R, Loppnau, P, Graslund, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Lovato, D.V, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-04-29 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of TgCDPK1 bound to NVPACU106
To Be Published
|
|
2BX4
 
 | |
2BX3
 
 | |
8DGG
 
 | | Structure of glycosylated LAG-3 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Silberstein, J.L, Mathews, I.I, Frank, J.A, Chan, K.-W, Fernandez, D, Du, J, Wang, J, Kong, X.-P, Cochran, J.R. | | Deposit date: | 2022-06-23 | | Release date: | 2022-08-17 | | Last modified: | 2022-08-31 | | Method: | X-RAY DIFFRACTION (3.78 Å) | | Cite: | Structural basis for LAG-3 dimeric association and inhibition of T cell function
To Be Published
|
|
6UT2
 
 | | 3D structure of the leiomodin/tropomyosin binding interface | | Descriptor: | Leiomodin-2, Tropomyosin alpha-1 chain chimeric peptide | | Authors: | Tolkatchev, D, Smith, G.E, Helms, G.L, Cort, J.R, Kostyukova, A.S. | | Deposit date: | 2019-10-29 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Leiomodin creates a leaky cap at the pointed end of actin-thin filaments.
Plos Biol., 18, 2020
|
|
1IZI
 
 | | Inhibitor of HIV protease with unusual binding mode potently inhibiting multi-resistant protease mutants | | Descriptor: | CHLORIDE ION, proteinase, {(1S)-1-BENZYL-4-[3-CARBAMOYL-1-(1-CARBAMOYL-2-PHENYL-ETHYLCARBAMOYL)-(S)-PROPYLCARBAMOYL]-2-OXO-5-PHENYL-PENTYL}-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Weber, J, Mesters, J.R, Lepsik, M, Prejdova, J, Svec, M, Sponarova, J, Mlcochova, P, Skalicka, K, Strisovsky, K, Uhlikova, T, Soucek, M, Machala, L, Stankova, M, Vondrasek, J, Klimkait, T, Kraeusslich, H.-G, Hilgenfeld, R, Konvalinka, J. | | Deposit date: | 2002-10-02 | | Release date: | 2002-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Unusual Binding Mode of an HIV-1 Protease Inhibitor Explains its Potency against Multi-drug-resistant Virus Strains
J.MOL.BIOL., 324, 2002
|
|
5JJW
 
 | | Crystal structure of the HAT domain of sart3 in complex with USP15 DUSP-UBL domain | | Descriptor: | 1,2-ETHANEDIOL, Squamous cell carcinoma antigen recognized by T-cells 3, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Zhang, Q, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-04-25 | | Release date: | 2016-05-04 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structure of the HAT domain of sart3 in complex with USP15 DUSP-UBL domain
to be published
|
|
2DN3
 
 | | 1.25A resolution crystal structure of human hemoglobin in the carbonmonoxy form | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha subunit, Hemoglobin beta subunit, ... | | Authors: | Park, S.-Y, Yokoyama, T, Shibayama, N, Shiro, Y, Tame, J.R. | | Deposit date: | 2006-04-25 | | Release date: | 2006-05-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25 a resolution crystal structures of human haemoglobin in the oxy, deoxy and carbonmonoxy forms.
J.Mol.Biol., 360, 2006
|
|
8CY5
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 39 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), N-{3-[1-(tert-butoxycarbonyl)piperidin-4-yl]propyl}adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXX
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 6 | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY3
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 15 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), N-[3-(4-aminophenyl)propyl]adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXS
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor MTA | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, DNA Strand 1, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
5J7D
 
 | | Computationally Designed Thioredoxin dF106 | | Descriptor: | COPPER (II) ION, Designed Thioredoxin dF106 | | Authors: | Horowitz, S, Johansen, N, Olsen, J.G, Winther, J.R. | | Deposit date: | 2016-04-06 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational Redesign of Thioredoxin Is Hypersensitive toward Minor Conformational Changes in the Backbone Template.
J.Mol.Biol., 428, 2016
|
|
8CXV
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 3 | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXT
 
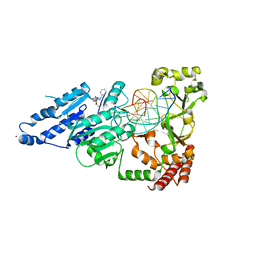 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor N6-benzyladenosine (Compound 1) | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY1
 
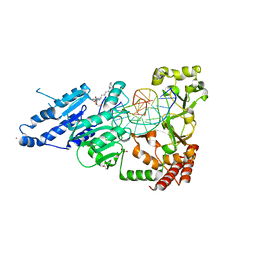 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 19 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(5-phenylpentyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXY
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor N6-(2-Phenethyl)adenosine (Compound 8) | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
6UYB
 
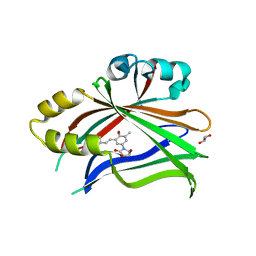 | | Crystal structure of TEAD2 bound to Compound 1 | | Descriptor: | (3R,4R)-1-{3-[(E)-2-(4-chlorophenyl)ethenyl]-4-methoxy-5-methylphenyl}-3,4-dihydroxypyrrolidin-2-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Noland, C.L, Holden, J.K, Crawford, J.J, Zbieg, J.R, Cunningham, C.N. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Small Molecule Dysregulation of TEAD Lipidation Induces a Dominant-Negative Inhibition of Hippo Pathway Signaling.
Cell Rep, 31, 2020
|
|
8CXZ
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor N6-(3-Phenylpropyl)adenosine (Compound 14) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(3-phenylpropyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
5IUT
 
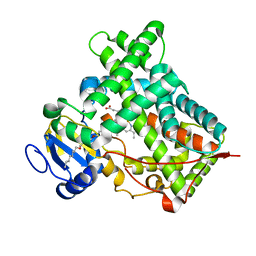 | | STRUCTURE OF P450 2B4 F202W MUTANT | | Descriptor: | 3,6,9,12,15,18-hexaoxahexacosan-1-ol, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jang, H.-H, Halpert, J.R, Shah, M.B. | | Deposit date: | 2016-03-18 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Effect of detergent binding on cytochrome P450 2B4 structure as analyzed by X-ray crystallography and deuterium-exchange mass spectrometry.
Biophys.Chem., 216, 2016
|
|
8CXU
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 2 | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY2
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor APNEA (Compound 9) | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY0
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor MC4756 (Compound 178) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(4-phenylbutyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXW
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor piclidenoson (Compound 4) | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
