4TN4
 
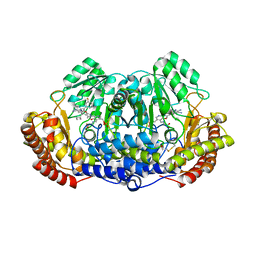 | | Crystal structure of ternary complex of Plasmodium vivax SHMT with glycine and a novel pyrazolopyran 33G: (4S)-6-amino-4-(5-cyano-3'-fluorobiphenyl-3-yl)-4-cyclobutyl-3-methyl-2,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile | | Descriptor: | (4S)-6-amino-4-(5-cyano-3'-fluorobiphenyl-3-yl)-4-cyclobutyl-3-methyl-2,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Witschel, M.C. | | Deposit date: | 2014-06-03 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of Plasmodial Serine Hydroxymethyltransferase (SHMT): Cocrystal Structures of Pyrazolopyrans with Potent Blood- and Liver-Stage Activities.
J.Med.Chem., 58, 2015
|
|
5WKJ
 
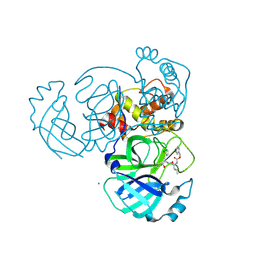 | | 2.05 A resolution structure of MERS 3CL protease in complex with inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, CALCIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-guided design of potent and permeable inhibitors of MERS coronavirus 3CL protease that utilize a piperidine moiety as a novel design element.
Eur J Med Chem, 150, 2018
|
|
5WKK
 
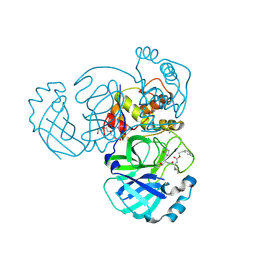 | | 1.55 A resolution structure of MERS 3CL protease in complex with inhibitor GC813 | | Descriptor: | (1R,2S)-2-[(N-{[2-(3-chlorophenyl)ethoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[2-(3-chlorophenyl)ethoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, MAGNESIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-guided design of potent and permeable inhibitors of MERS coronavirus 3CL protease that utilize a piperidine moiety as a novel design element.
Eur J Med Chem, 150, 2018
|
|
5WKM
 
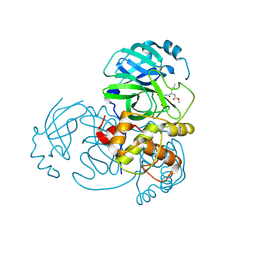 | | 2.25 A resolution structure of MERS 3CL protease in complex with piperidine-based peptidomimetic inhibitor 21 | | Descriptor: | (1R,2S)-2-{[N-({[1-(tert-butoxycarbonyl)-4-ethylpiperidin-4-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[1-(tert-butoxycarbonyl)-4-ethylpiperidin-4-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-guided design of potent and permeable inhibitors of MERS coronavirus 3CL protease that utilize a piperidine moiety as a novel design element.
Eur J Med Chem, 150, 2018
|
|
4O7I
 
 | | Structural and functional characterization of 3'(2'),5'-bisphosphate nucleotidase1 from Entamoeba histolytica | | Descriptor: | 3'(2'),5'-bisphosphate nucleotidase, putative, MAGNESIUM ION, ... | | Authors: | Tarique, K.F, Rehman, S.A.A, Gourinath, S. | | Deposit date: | 2013-12-24 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural elucidation of a dual-activity PAP phosphatase-1 from Entamoeba histolytica capable of hydrolysing both 3'-phosphoadenosine 5'-phosphate and inositol 1,4-bisphosphate
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4WJS
 
 | |
6SWV
 
 | | Trypsin fast data collection | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | von Stetten, D, Mueller-Dieckmann, C. | | Deposit date: | 2019-09-24 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | ID30A-3 (MASSIF-3) - a beamline for macromolecular crystallography at the ESRF with a small intense beam.
J.Synchrotron Radiat., 27, 2020
|
|
2HY3
 
 | | Crystal structure of the human tyrosine receptor phosphate gamma in complex with vanadate | | Descriptor: | Receptor-type tyrosine-protein phosphatase gamma, VANADATE ION | | Authors: | Jin, X, Min, T, Bera, A, Mu, H, Sauder, J.M, Freeman, J.C, Reyes, C, Smith, D, Wasserman, S.R, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
4WJV
 
 | | Crystal structure of Rsa4 in complex with the Nsa2 binding peptide | | Descriptor: | Maltose-binding periplasmic protein, Ribosome assembly protein 4, Ribosome biogenesis protein NSA2, ... | | Authors: | Holdermann, I, Paternoga, H, Bassler, J, Hurt, E, Sinning, I. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
4WJU
 
 | | Crystal structure of Rsa4 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Ribosome assembly protein 4 | | Authors: | Holdermann, I, Bassler, J, Hurt, E, Sinning, I. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
1XUC
 
 | | Matrix metalloproteinase-13 complexed with non-zinc binding inhibitor | | Descriptor: | CALCIUM ION, Collagenase 3, N,N'-BIS(3-METHYLBENZYL)PYRIMIDINE-4,6-DICARBOXAMIDE, ... | | Authors: | Engel, C.K, Wendt, K.U. | | Deposit date: | 2004-10-26 | | Release date: | 2005-10-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the highly selective inhibition of MMP-13
Chem.Biol., 12, 2005
|
|
2PMQ
 
 | | Crystal structure of a mandelate racemase/muconate lactonizing enzyme from Roseovarius sp. HTCC2601 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Lau, C, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of new enzymes and metabolic pathways by using structure and genome context.
Nature, 502, 2013
|
|
3M0E
 
 | | Crystal structure of the ATP-bound state of Walker B mutant of NtrC1 ATPase domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Transcriptional regulator (NtrC family) | | Authors: | Chen, B, Sysoeva, T.A, Chowdhury, S, Rusu, M, Birmanns, S, Guo, L, Hanson, J, Yang, H, Nixon, B.T. | | Deposit date: | 2010-03-02 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Engagement of Arginine Finger to ATP Triggers Large Conformational Changes in NtrC1 AAA+ ATPase for Remodeling Bacterial RNA Polymerase.
Structure, 18, 2010
|
|
1XUD
 
 | | Matrix metalloproteinase-13 complexed with non-zinc binding inhibitor | | Descriptor: | CALCIUM ION, Collagenase 3, N,N'-BIS(4-FLUORO-3-METHYLBENZYL)PYRIMIDINE-4,6-DICARBOXAMIDE, ... | | Authors: | Engel, C.K, Wendt, K.U. | | Deposit date: | 2004-10-26 | | Release date: | 2005-10-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the highly selective inhibition of MMP-13.
Chem.Biol., 12, 2005
|
|
1XUR
 
 | | Matrix metalloproteinase-13 complexed with non-zinc binding inhibitor | | Descriptor: | CALCIUM ION, Collagenase 3, N,N'-BIS(PYRIDIN-3-YLMETHYL)PYRIMIDINE-4,6-DICARBOXAMIDE, ... | | Authors: | Engel, C.K, Wendt, K.U. | | Deposit date: | 2004-10-26 | | Release date: | 2005-10-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the highly selective inhibition of MMP-13.
Chem.Biol., 12, 2005
|
|
3TJP
 
 | | Crystal Structure of PI3K gamma with N6-(3,4-dimethoxyphenyl)-2-morpholino-[4,5'-bipyrimidine]-2',6-diamine | | Descriptor: | N~6~-(3,4-dimethoxyphenyl)-2-(morpholin-4-yl)-4,5'-bipyrimidine-2',6-diamine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Knapp, M.S, Elling, R.A, Ornelas, E. | | Deposit date: | 2011-08-24 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Identification of 5-(2,4-dimorpholinopyrimidin-6-yl)-4-(trifluoromethyl)pyridin-2-amine (NVP-BKM120) as a Potent, Selective and Orally Bioavailable Class I PI3 Kinase Inhibitor for the Treatment of Cancer
To be Published
|
|
4ZPL
 
 | | Crystal Structure of Protocadherin Beta 1 EC1-3 | | Descriptor: | CALCIUM ION, Protein Pcdhb1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Goodman, K.M, Bahna, F, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
2HHL
 
 | | Crystal structure of the human small CTD phosphatase 3 isoform 1 | | Descriptor: | 12-TUNGSTOPHOSPHATE, CTD small phosphatase-like protein | | Authors: | Malashkevich, V.N, Toro, R, Ramagopal, U, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-28 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2L0X
 
 | | Solution structure of the 21 kDa GTPase RHEB bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Stoll, R, Heumann, R, Berghaus, C, Kock, G. | | Deposit date: | 2010-07-19 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Ras homolog enriched in brain (Rheb) enhances apoptotic signaling.
J.Biol.Chem., 285, 2010
|
|
3N7C
 
 | | Crystal structure of the RAN binding domain from the nuclear pore complex component NUP2 from Ashbya gossypii | | Descriptor: | ABR034Wp | | Authors: | Sampathkumar, P, Manglicmot, D, Gilmore, J, Bain, K, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-26 | | Release date: | 2010-06-16 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of the RAN binding domain from the nuclear pore complex component NUP2 from Ashbya gossypii
To be Published
|
|
4XX1
 
 | | Low resolution structure of LCAT in complex with Fab1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1 heavy chain, Fab1 light chain, ... | | Authors: | Piper, D.E, Walker, N.P.C, Romanow, W.G, Thibault, S.T. | | Deposit date: | 2015-01-29 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The high-resolution crystal structure of human LCAT.
J.Lipid Res., 56, 2015
|
|
3AMV
 
 | | ALLOSTERIC INHIBITION OF GLYCOGEN PHOSPHORYLASE A BY A POTENTIAL ANTIDIABETIC DRUG | | Descriptor: | 2,3-DICARBOXY-4-(2-CHLORO-PHENYL)-1-ETHYL-5-ISOPROPOXYCARBONYL-6-METHYL-PYRIDINIUM, GLYCEROL, PROTEIN (GLYCOGEN PHOSPHORYLASE), ... | | Authors: | Oikonomakos, N.G, Tsitsanou, K.E, Zographos, S.E, Skamnaki, V.T. | | Deposit date: | 1999-02-18 | | Release date: | 1999-02-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allosteric inhibition of glycogen phosphorylase a by the potential antidiabetic drug 3-isopropyl 4-(2-chlorophenyl)-1,4-dihydro-1-ethyl-2-methyl-pyridine-3,5,6-tricarbo xylate.
Protein Sci., 8, 1999
|
|
3O57
 
 | | Catalytic domain of human phosphodiesterase 4b2b in complex with a 5-heterocycle pyrazolopyridine inhibitor | | Descriptor: | 5-[5-benzyl-4-(2-oxo-2-pyrrolidin-1-ylethyl)-1,3-oxazol-2-yl]-1-ethyl-N-(tetrahydro-2H-pyran-4-yl)-1H-pyrazolo[3,4-b]pyridin-4-amine, ARSENIC, GLYCEROL, ... | | Authors: | Somers, D.O, Neu, M. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyrazolopyridines as potent PDE4B inhibitors: 5-heterocycle SAR.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4XWG
 
 | | Crystal Structure of LCAT (C31Y) in complex with Fab1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1 Heavy Chain, Fab1 Light Chain, ... | | Authors: | Piper, D.E, Walker, N.P.C, Romanow, W.G, Thibault, S.T. | | Deposit date: | 2015-01-28 | | Release date: | 2015-07-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The high-resolution crystal structure of human LCAT.
J.Lipid Res., 56, 2015
|
|
2YHI
 
 | | Trypanosoma brucei PTR1 in complex with inhibitor WH16 | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Nerini, E, Dawson, A, Hunter, W.N, Costi, M.P. | | Deposit date: | 2011-05-03 | | Release date: | 2012-05-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploiting the 2-Amino-1,3,4-thiadiazole Scaffold To Inhibit Trypanosoma brucei Pteridine Reductase in Support of Early-Stage Drug Discovery.
ACS Omega, 2, 2017
|
|
