1KF4
 
 | | Atomic Resolution Structure of RNase A at pH 6.3 | | Descriptor: | SULFATE ION, pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KF8
 
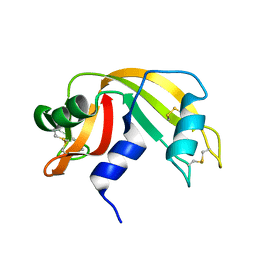 | | Atomic resolution structure of RNase A at pH 8.8 | | Descriptor: | pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2C8J
 
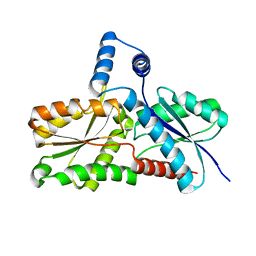 | | CRYSTAL STRUCTURE OF ferrochelatase HemH-1 from Bacillus anthracis, str. Ames | | Descriptor: | FERROCHELATASE 1 | | Authors: | Muller, A, Lebedev, A.A, Moroz, O.V, Blagova, E.V, Levdikov, V.M, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2005-12-05 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Ferrochelatase Hemh-1 from Bacillus Anthracis, Str. Ames
To be Published
|
|
1KF3
 
 | | Atomic Resolution Structure of RNase A at pH 5.9 | | Descriptor: | SULFATE ION, pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2CEY
 
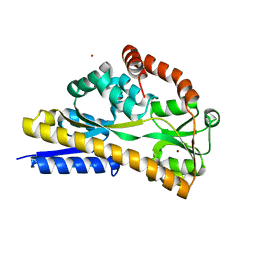 | | Apo Structure of SiaP | | Descriptor: | PROTEIN HI0146, ZINC ION | | Authors: | Muller, A, Severi, E, Mulligan, C, Watts, A.G, Kelly, D.J, Wilson, K.S, Wilkinson, A.J, Thomas, G.H. | | Deposit date: | 2006-02-11 | | Release date: | 2006-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conservation of Structure and Mechanism in Primary and Secondary Transporters Exemplified by Siap, a Sialic Acid Binding Virulence Factor from Haemophilus Influenzae
J.Biol.Chem., 281, 2006
|
|
1KF5
 
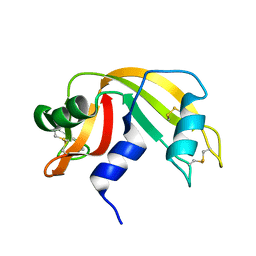 | | Atomic Resolution Structure of RNase A at pH 7.1 | | Descriptor: | pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2CTC
 
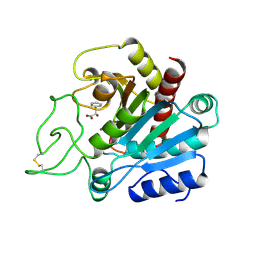 | | THE HIGH RESOLUTION CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN CARBOXYPEPTIDASE A AND L-PHENYL LACTATE | | Descriptor: | ALPHA-HYDROXY-BETA-PHENYL-PROPIONIC ACID, CARBOXYPEPTIDASE A, ZINC ION | | Authors: | Teplyakov, A, Wilson, K.S, Orioli, P, Mangani, S. | | Deposit date: | 1993-04-02 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structure of the complex between carboxypeptidase A and L-phenyl lactate.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1KF2
 
 | | Atomic Resolution Structure of RNase A at pH 5.2 | | Descriptor: | SULFATE ION, pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2FBA
 
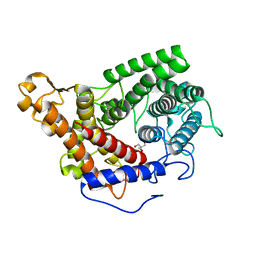 | | Glucoamylase from Saccharomycopsis fibuligera at atomic resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glucoamylase GLU1 | | Authors: | Sevcik, J, Hostinova, E, Solovicova, A, Gasperik, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 2005-12-09 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of the complex of a yeast glucoamylase with acarbose reveals the presence of a raw starch binding site on the catalytic domain.
Febs J., 273, 2006
|
|
2CEX
 
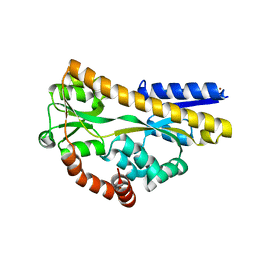 | | Structure of a sialic acid binding protein (SiaP) in the presence of the sialic acid acid analogue Neu5Ac2en | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, PROTEIN HI0146, ... | | Authors: | Muller, A, Severi, E, Mulligan, C, Watts, A.G, Kelly, D.J, Wilson, K.S, Wilkinson, A.J, Thomas, G.H. | | Deposit date: | 2006-02-10 | | Release date: | 2006-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conservation of Structure and Mechanism in Primary and Secondary Transporters Exemplified by Siap, a Sialic Acid Binding Virulence Factor from Haemophilus Influenzae
J.Biol.Chem., 281, 2006
|
|
2CTX
 
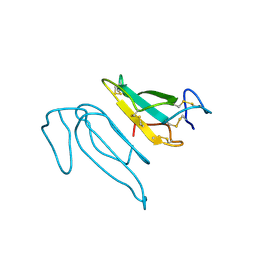 | | THE REFINED CRYSTAL STRUCTURE OF ALPHA-COBRATOXIN FROM NAJA NAJA SIAMENSIS AT 2.4-ANGSTROMS RESOLUTION | | Descriptor: | ALPHA-COBRATOXIN | | Authors: | Betzel, C, Lange, G, Pal, G.P, Wilson, K.S, Maelicke, A, Saenger, W. | | Deposit date: | 1991-09-24 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The refined crystal structure of alpha-cobratoxin from Naja naja siamensis at 2.4-A resolution.
J.Biol.Chem., 266, 1991
|
|
2FDN
 
 | | 2[4FE-4S] FERREDOXIN FROM CLOSTRIDIUM ACIDI-URICI | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Dauter, Z, Wilson, K.S, Sieker, L.C, Meyer, J, Moulis, J.M. | | Deposit date: | 1997-10-01 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Atomic resolution (0.94 A) structure of Clostridium acidurici ferredoxin. Detailed geometry of [4Fe-4S] clusters in a protein.
Biochemistry, 36, 1997
|
|
5A5V
 
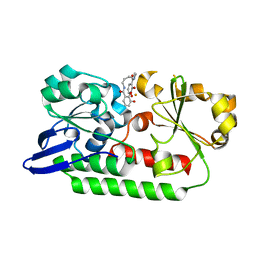 | | A complex of the synthetic siderophore analogue Fe(III)-6-LICAM with the CeuE periplasmic protein from Campylobacter jejuni | | Descriptor: | ENTEROCHELIN UPTAKE PERIPLASMIC BINDING PROTEIN, FE (III) ION, N,N'-hexane-1,4-diylbis(2,3-dihydroxybenzamide) | | Authors: | Blagova, E, Hughes, A, Moroz, O.V, Raines, D.J, Wilde, E.J, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2015-06-22 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
5AP9
 
 | | Controlled lid-opening in Thermomyces lanuginosus lipase - a switch for activity and binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Skjold-Joergensen, J, Vind, J, Moroz, O.V, Blagova, E.V, Bhatia, V.K, Svendsen, A, Wilson, K.S, Bjerrum, M.J. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Controlled lid-opening in Thermomyces lanuginosus lipase- An engineered switch for studying lipase function.
Biochim. Biophys. Acta, 1865, 2017
|
|
2TPL
 
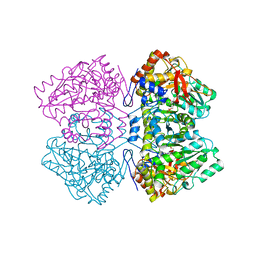 | | TYROSINE PHENOL-LYASE FROM CITROBACTER INTERMEDIUS COMPLEX WITH 3-(4'-HYDROXYPHENYL)PROPIONIC ACID, PYRIDOXAL-5'-PHOSPHATE AND CS+ ION | | Descriptor: | CESIUM ION, HYDROXYPHENYL PROPIONIC ACID, TYROSINE PHENOL-LYASE | | Authors: | Antson, A.A, Demidkina, T.V, Wilson, K.S. | | Deposit date: | 1997-01-23 | | Release date: | 1997-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of Citrobacter freundii tyrosine phenol-lyase complexed with 3-(4'-hydroxyphenyl)propionic acid, together with site-directed mutagenesis and kinetic analysis, demonstrates that arginine 381 is required for substrate specificity.
Biochemistry, 36, 1997
|
|
2TEC
 
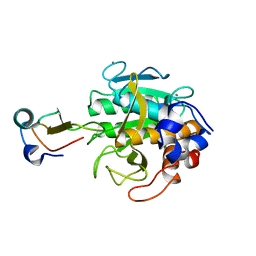 | | MOLECULAR DYNAMICS REFINEMENT OF A THERMITASE-EGLIN-C COMPLEX AT 1.98 ANGSTROMS RESOLUTION AND COMPARISON OF TWO CRYSTAL FORMS THAT DIFFER IN CALCIUM CONTENT | | Descriptor: | CALCIUM ION, EGLIN C, THERMITASE | | Authors: | Gros, P, Betzel, C, Dauter, Z, Wilson, K.S, Hol, W.G.J. | | Deposit date: | 1990-10-26 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular dynamics refinement of a thermitase-eglin-c complex at 1.98 A resolution and comparison of two crystal forms that differ in calcium content.
J.Mol.Biol., 210, 1989
|
|
2RVE
 
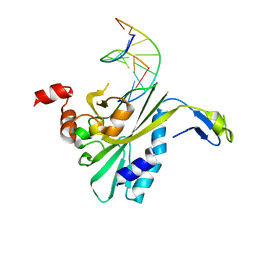 | | THE CRYSTAL STRUCTURE OF ECORV ENDONUCLEASE AND OF ITS COMPLEXES WITH COGNATE AND NON-COGNATE DNA SEGMENTS | | Descriptor: | DNA (5'-D(*CP*GP*AP*GP*CP*TP*CP*G)-3'), PROTEIN (ECO RV (E.C.3.1.21.4)) | | Authors: | Winkler, F.K, Banner, D.W, Oefner, C, Tsernoglou, D, Brown, R.S, Heathman, S.P, Bryan, R.K, Martin, P.D, Petratos, K, Wilson, K.S. | | Deposit date: | 1991-03-19 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments.
EMBO J., 12, 1993
|
|
2UVK
 
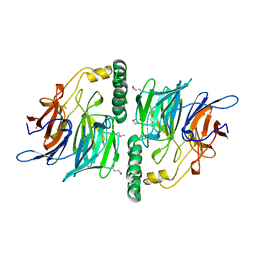 | | Structure of YjhT | | Descriptor: | YJHT | | Authors: | Muller, A, Severi, E, Wilson, K.S, Thomas, G.H. | | Deposit date: | 2007-03-12 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sialic Acid Mutarotation is Catalyzed by the Escherichia Coli Beta-Propeller Protein Yjht.
J.Biol.Chem., 283, 2008
|
|
2WHL
 
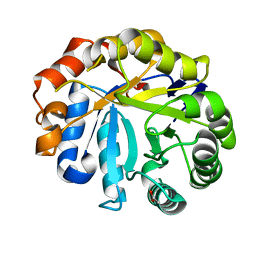 | | Understanding how diverse mannanases recognise heterogeneous substrates | | Descriptor: | ACETATE ION, BETA-MANNANASE, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Tailford, L.E, Ducros, V.M.A, Flint, J.E, Roberts, S.M, Morland, C, Zechel, D.L, Smith, N, Bjornvad, M.E, Borchert, T.V, Wilson, K.S, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2009-05-05 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Understanding How Diverse -Mannanases Recognise Heterogeneous Substrates.
Biochemistry, 48, 2009
|
|
2W6L
 
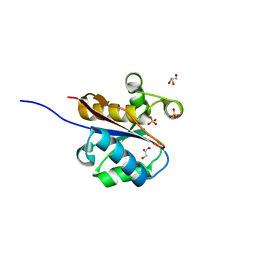 | | The crystal structure at 1.7 A resolution of CobE, a protein from the cobalamin (vitamin B12) biosynthetic pathway | | Descriptor: | COBE, GLYCEROL, SULFATE ION | | Authors: | Vevodova, J, Smith, D, McGoldrick, H, Deery, E, Murzin, A.G, Warren, M.J, Wilson, K.S. | | Deposit date: | 2008-12-18 | | Release date: | 2008-12-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Crystal Structure at 1.7 A Resolution of Cobe, a Protein from the Cobalamin (Vitamin B12) Biosynthetic Pathway
To be Published
|
|
2WAN
 
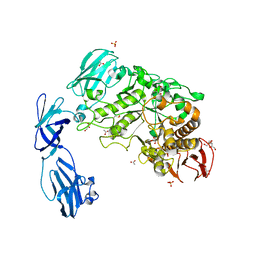 | | Pullulanase from Bacillus acidopullulyticus | | Descriptor: | ACETATE ION, GLYCEROL, PULLULANASE, ... | | Authors: | Turkenburg, J.P, Brzozowski, A.M, Svendsen, A, Borchert, T.V, Davies, G.J, Wilson, K.S. | | Deposit date: | 2009-02-10 | | Release date: | 2009-05-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of a Pullulanase from Bacillus Acidopullulyticus.
Proteins, 76, 2009
|
|
2X8J
 
 | | Intracellular subtilisin precursor from B. clausii | | Descriptor: | GLYCEROL, INTRACELLULAR SUBTILISIN PROTEASE, PENTAETHYLENE GLYCOL, ... | | Authors: | Vedodova, J, Gamble, M, Ariza, A, Dodson, E, Jones, D.D, Wilson, K.S. | | Deposit date: | 2010-03-09 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of an intracellular subtilisin reveals novel structural features unique to this subtilisin family.
Structure, 18, 2010
|
|
5ADW
 
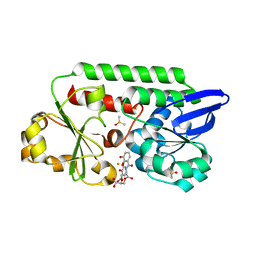 | | The Periplasmic Binding Protein CeuE of Campylobacter jejuni preferentially binds the iron(III) complex of the Linear Dimer Component of Enterobactin | | Descriptor: | 2S-2-[(2,3-DIHYDROXYPHENYL)CARBONYLAMINO]-3-[(2S)-2-[(2,3-DIHYDROXYPHENYL)CARBONYLAMINO]-3-HYDROXY-PROPANOYL]OXY-PROPANOIC ACID, DIMETHYL SULFOXIDE, ENTEROCHELIN UPTAKE PERIPLASMIC BINDING PROTEIN, ... | | Authors: | Raines, D.J, Moroz, O.V, Turkenburg, J.P, Wilson, K.S, Duhme-Klair, A.K. | | Deposit date: | 2015-08-24 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacteria in an Intense Competition for Iron: Key Component of the Campylobacter Jejuni Iron Uptake System Scavenges Enterobactin Hydrolysis Product.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2WWG
 
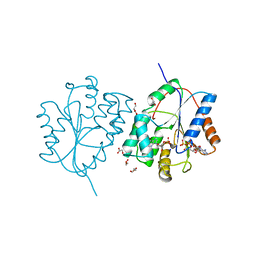 | | Plasmodium falciparum thymidylate kinase in complex with dGMP and ADP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Whittingham, J.L, Carrero-Lerida, J, Brannigan, J.A, Ruiz-Perez, L.M, Silva, A.P, Fogg, M.J, Wilkinson, A.J, Gilbert, I.H, Wilson, K.S, Gonzalez-Pacanowska, D. | | Deposit date: | 2009-10-23 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Efficient Phosphorylation of Aztmp and Dgmp by Plasmodium Falciparum Type I Thymidylate Kinase.
Biochem.J., 428, 2010
|
|
2WHJ
 
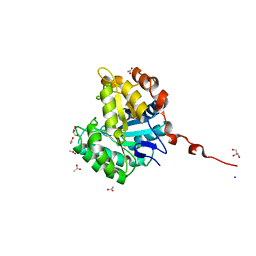 | | Understanding how diverse mannanases recognise heterogeneous substrates | | Descriptor: | ACETATE ION, BETA-MANNANASE, GLYCEROL, ... | | Authors: | Tailford, L.E, Ducros, V.M.A, Flint, J.E, Roberts, S.M, Morland, C, Zechel, D.L, Smith, N, Bjornvad, M.E, Borchert, T.V, Wilson, K.S, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2009-05-05 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Understanding How Diverse -Mannanases Recognise Heterogeneous Substrates.
Biochemistry, 48, 2009
|
|
