8H0D
 
 | | Structure of the thermolabile hemolysin from Vibrio alginolyticus (in complex with docosahexaenoic acid) | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, MAGNESIUM ION, ... | | Authors: | Ma, Q, Wang, C. | | Deposit date: | 2022-09-28 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Catalytic site flexibility facilitates the substrate and catalytic promiscuity of Vibrio dual lipase/transferase.
Nat Commun, 14, 2023
|
|
5F98
 
 | | Crystal structure of RIG-I in complex with Cap-0 RNA | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Probable ATP-dependent RNA helicase DDX58, ... | | Authors: | Wang, C, Marcotrigiano, J, Miller, M, Jiang, F. | | Deposit date: | 2015-12-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8H09
 
 | |
5F9H
 
 | | Crystal structure of RIG-I helicase-RD in complex with 24-mer 5' triphosphate hairpin RNA | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable ATP-dependent RNA helicase DDX58, ... | | Authors: | Wang, C, Marcotrigiano, J, Miller, M, Jiang, F. | | Deposit date: | 2015-12-09 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5HJK
 
 | |
5F9F
 
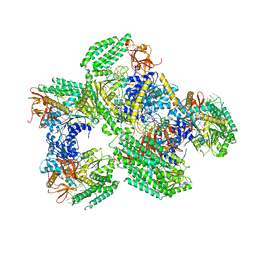 | | Crystal structure of RIG-I helicase-RD in complex with 24-mer blunt-end hairpin RNA | | Descriptor: | (R,R)-2,3-BUTANEDIOL, MAGNESIUM ION, Probable ATP-dependent RNA helicase DDX58, ... | | Authors: | Wang, C, Marcotrigiano, J, Miller, M.T, Jiang, F. | | Deposit date: | 2015-12-09 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4LWP
 
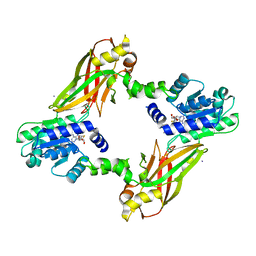 | | Crystal structure of PRMT6-SAH | | Descriptor: | Arginine N-methyltransferase, putative, IODIDE ION, ... | | Authors: | Zhu, Y, Wang, C, Shi, Y, Teng, M. | | Deposit date: | 2013-07-28 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Crystal Structure of Arginine Methyltransferase 6 from Trypanosoma brucei
Plos One, 9, 2014
|
|
4LWO
 
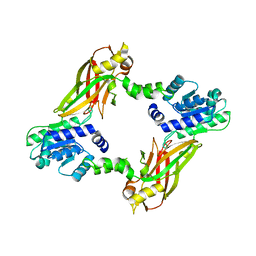 | | Crystal structure of PRMT6 | | Descriptor: | Arginine N-methyltransferase, putative | | Authors: | Zhu, Y, Wang, C, Shi, Y, Teng, M. | | Deposit date: | 2013-07-28 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal Structure of Arginine Methyltransferase 6 from Trypanosoma brucei
Plos One, 9, 2014
|
|
4DCK
 
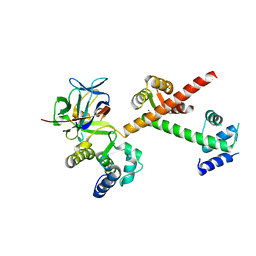 | | Crystal structure of the C-terminus of voltage-gated sodium channel in complex with FGF13 and CaM | | Descriptor: | Calmodulin, Fibroblast growth factor 13, MAGNESIUM ION, ... | | Authors: | Chung, B.C, Wang, C, Yan, H, Pitt, G.S, Lee, S.Y. | | Deposit date: | 2012-01-17 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Ternary Complex of a NaV C-Terminal Domain, a Fibroblast Growth Factor Homologous Factor, and Calmodulin.
Structure, 20, 2012
|
|
5WT3
 
 | |
5WT1
 
 | |
4O8B
 
 | | Crystal structure of transcriptional regulator BswR | | Descriptor: | Uncharacterized protein | | Authors: | Ye, F.Z, Wang, C, Kumar, V, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2013-12-26 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | BswR controls bacterial motility and biofilm formation in Pseudomonas aeruginosa through modulation of the small RNA rsmZ.
Nucleic Acids Res., 42, 2014
|
|
4OF9
 
 | | Structure of K42N variant of sperm whale myoglobin | | Descriptor: | 1,2-ETHANEDIOL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lebioda, L, Wang, C, Lovelace, L.L. | | Deposit date: | 2014-01-14 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.241 Å) | | Cite: | Structures of K42N and K42Y sperm whale myoglobins point to an inhibitory role of distal water in peroxidase activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5XOE
 
 | |
4OOD
 
 | | Structure of K42Y mutant of sperm whale myoglobin | | Descriptor: | 1,2-ETHANEDIOL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lebioda, L, Wang, C, Lovelace, L.L. | | Deposit date: | 2014-01-31 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structures of K42N and K42Y sperm whale myoglobins point to an inhibitory role of distal water in peroxidase activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4DC2
 
 | | Structure of PKC in Complex with a Substrate Peptide from Par-3 | | Descriptor: | ADENINE, Partitioning defective 3 homolog, Protein kinase C iota type | | Authors: | Shang, Y, Wang, C, Yu, J, Zhang, M. | | Deposit date: | 2012-01-17 | | Release date: | 2012-07-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Substrate recognition mechanism of atypical protein kinase Cs revealed by the structure of PKC iota in complex with a substrate peptide from Par-3
Structure, 20, 2012
|
|
4KJT
 
 | | Structure of the L100F MUTANT OF DEHALOPEROXIDASE-HEMOGLOBIN A FROM AMPHITRITE ORNATA WITH OXYGEN | | Descriptor: | 1,2-ETHANEDIOL, Dehaloperoxidase A, OXYGEN MOLECULE, ... | | Authors: | Wang, C, Lovelace, L, Lebioda, L. | | Deposit date: | 2013-05-03 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Influence of heme environment structure on dioxygen affinity for the dual function Amphitrite ornata hemoglobin/dehaloperoxidase. Insights into the evolutional structure-function adaptations.
Arch.Biochem.Biophys., 545, 2014
|
|
7D4B
 
 | | Crystal structure of 4-1BB in complex with a VHH | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wang, C. | | Deposit date: | 2020-09-23 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Generation of a safe and efficacious llama single-domain antibody fragment (vHH) targeting the membrane-proximal region of 4-1BB for engineering therapeutic bispecific antibodies for cancer.
J Immunother Cancer, 9, 2021
|
|
7CZD
 
 | | Crystal structure of PD-L1 in complex with a VHH | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Programmed cell death 1 ligand 1, ... | | Authors: | Wang, C. | | Deposit date: | 2020-09-08 | | Release date: | 2021-07-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Generation of a safe and efficacious llama single-domain antibody fragment (vHH) targeting the membrane-proximal region of 4-1BB for engineering therapeutic bispecific antibodies for cancer.
J Immunother Cancer, 9, 2021
|
|
6A9X
 
 | | Crystal Structure of AnkG/GABARAP Complex | | Descriptor: | Ankyrin-3, Gamma-aminobutyric acid receptor-associated protein | | Authors: | Wang, C, Li, J, Chen, K, Zhang, M. | | Deposit date: | 2018-07-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Ankyrin-G regulates forebrain connectivity and network synchronization via interaction with GABARAP.
Mol. Psychiatry, 2018
|
|
6VZ3
 
 | | Escherichia coli transcription-translation complex D2 (TTC-D2) containing mRNA with a 27 nt long spacer | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6VU3
 
 | | Cryo-EM structure of Escherichia coli transcription-translation complex A (TTC-A) containing mRNA with a 12 nt long spacer | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R. | | Deposit date: | 2020-02-14 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6VZJ
 
 | | Escherichia coli transcription-translation complex A1 (TTC-A1) containing mRNA with a 15 nt long spacer, fMet-tRNAs at E-site and P-site, and lacking transcription factor NusG | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-28 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6VZ5
 
 | | Escherichia coli transcription-translation complex D3 (TTC-D3) containing mRNA with a 21 nt long spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6VYZ
 
 | | Escherichia coli transcription-translation complex C6 (TTC-C6) containing mRNA with a 21 nt long spacer, NusA, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
