5XFE
 
 | | Luciferin-regenerating enzyme solved by SAD using XFEL (refined against 11,000 patterns) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2017-04-10 | | Release date: | 2017-08-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental phase determination with selenomethionine or mercury-derivatization in serial femtosecond crystallography
IUCrJ, 4, 2017
|
|
1FXZ
 
 | | CRYSTAL STRUCTURE OF SOYBEAN PROGLYCININ A1AB1B HOMOTRIMER | | Descriptor: | GLYCININ G1 | | Authors: | Adachi, M, Takenaka, Y, Gidamis, A.B, Mikami, B, Utsumi, S. | | Deposit date: | 2000-09-28 | | Release date: | 2001-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of soybean proglycinin A1aB1b homotrimer.
J.Mol.Biol., 305, 2001
|
|
7FG2
 
 | |
7FG7
 
 | |
7FG3
 
 | |
5XFC
 
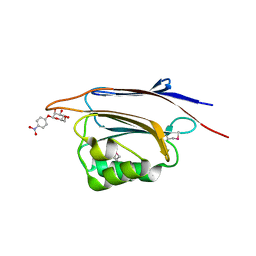 | | Serial femtosecond X-ray structure of a stem domain of human O-mannose beta-1,2-N-acetylglucosaminyltransferase solved by Se-SAD using XFEL (refined against 13,000 patterns) | | Descriptor: | 4-nitrophenyl beta-D-mannopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Fumiaki, Y, Kato, R, Manya, H. | | Deposit date: | 2017-04-10 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Experimental phase determination with selenomethionine or mercury-derivatization in serial femtosecond crystallography
IUCrJ, 4, 2017
|
|
7K8L
 
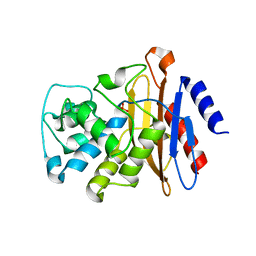 | | Beta-lactamase, Unmixed | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8000102 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7K8E
 
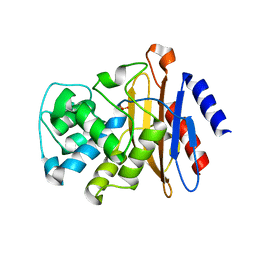 | | Beta-lactamase mixed with Ceftriaxone, 5ms | | Descriptor: | Beta-lactamase, Ceftriaxone, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-26 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.40005636 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7K8H
 
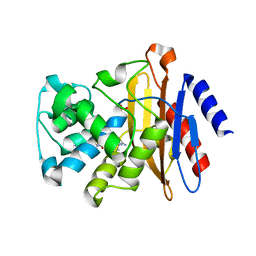 | | Beta-lactamase mixed with Ceftriaxone, 50ms | | Descriptor: | Beta-lactamase, Ceftriaxone, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.60006261 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7K8K
 
 | | Beta-lactamase mixed with Sulbactam, 60ms | | Descriptor: | Beta-lactamase, PHOSPHATE ION, SULBACTAM, ... | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7K8F
 
 | | Beta-lactamase mixed with Ceftriaxone, 10ms | | Descriptor: | Beta-lactamase, Ceftriaxone, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-26 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.60003138 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7C3G
 
 | | Crystal structure of human ALK2 kinase domain with R206H mutation in complex with a bicyclic pyrazole inhibitor RK-73134 | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type-1, SULFATE ION, ... | | Authors: | Sakai, N, Mishima-Tsumagari, C, Matsumoto, T, Shirouzu, M. | | Deposit date: | 2020-05-12 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Novel bicyclic pyrazoles as potent ALK2 (R206H) inhibitors for the treatment of fibrodysplasia ossificans progressiva.
Bioorg.Med.Chem.Lett., 38, 2021
|
|
7DEX
 
 | | Crystal Structures of Anthocyanin 5,3'-aromatic acyltransferase H174A mutant with caffeoyl-CoA | | Descriptor: | Anthocyanin 5-aromatic acyltransferase, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (E)-3-[3,4-bis(oxidanyl)phenyl]prop-2-enethioate | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2020-11-05 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora, a structural insight into biosynthesis of a blue anthocyanin.
Phytochemistry, 186, 2021
|
|
7DEV
 
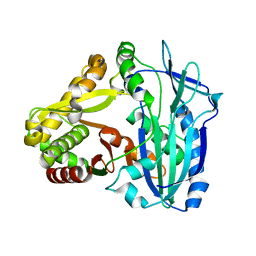 | | Crystal Structures of Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora | | Descriptor: | Anthocyanin 5-aromatic acyltransferase | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2020-11-05 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora, a structural insight into biosynthesis of a blue anthocyanin.
Phytochemistry, 186, 2021
|
|
2SFA
 
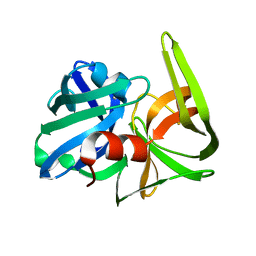 | | SERINE PROTEINASE FROM STREPTOMYCES FRADIAE ATCC 14544 | | Descriptor: | SERINE PROTEINASE | | Authors: | Kitadokoro, K, Tsuzuki, H. | | Deposit date: | 1994-04-25 | | Release date: | 1996-06-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Purification, characterization, primary structure, crystallization and preliminary crystallographic study of a serine proteinase from Streptomyces fradiae ATCC 14544.
Eur.J.Biochem., 220, 1994
|
|
1EWT
 
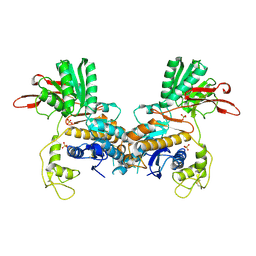 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 LIGAND FREE FORM I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1, SULFATE ION | | Authors: | Kunishima, N, Shimada, Y, Tsuji, Y, Jingami, H, Morikawa, K. | | Deposit date: | 2000-04-27 | | Release date: | 2000-12-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
1EWV
 
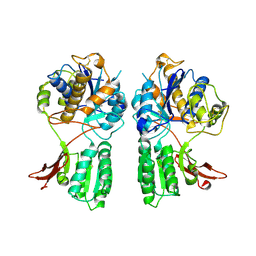 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 LIGAND FREE FORM II | | Descriptor: | METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 | | Authors: | Kunishima, N, Shimada, Y, Tsuji, Y, Jingami, H, Morikawa, K. | | Deposit date: | 2000-04-27 | | Release date: | 2000-12-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
1EWK
 
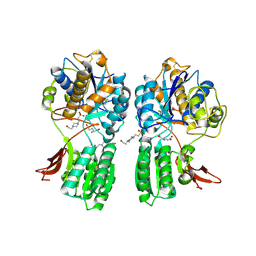 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 COMPLEXED WITH GLUTAMATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLUTAMIC ACID, ... | | Authors: | Kunishima, N, Shimada, Y, Jingami, H, Morikawa, K. | | Deposit date: | 2000-04-26 | | Release date: | 2000-12-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
5YO8
 
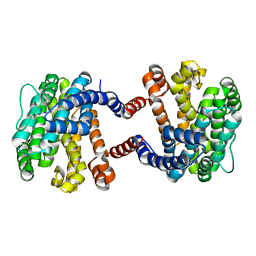 | |
1C8N
 
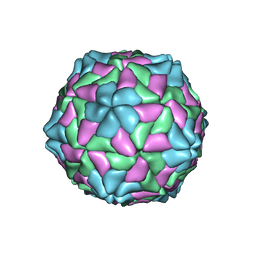 | | TOBACCO NECROSIS VIRUS | | Descriptor: | CALCIUM ION, COAT PROTEIN | | Authors: | Oda, Y, Fukuyama, K. | | Deposit date: | 2000-05-20 | | Release date: | 2000-08-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of tobacco necrosis virus at 2.25 A resolution.
J.Mol.Biol., 300, 2000
|
|
1IOP
 
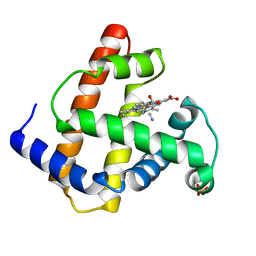 | | INCORPORATION OF A HEMIN WITH THE SHORTEST ACID SIDE-CHAINS INTO MYOGLOBIN | | Descriptor: | 6,7-DICARBOXYL-1,2,3,4,5,8-HEXAMETHYLHEMIN, CYANIDE ION, MYOGLOBIN, ... | | Authors: | Igarashi, N, Neya, S, Funasaki, N, Tanaka, N. | | Deposit date: | 1997-12-12 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of 6,7-dicarboxyheme-substituted myoglobin
Biochemistry, 37, 1998
|
|
7E4L
 
 | |
3WQP
 
 | | Crystal structure of Rubisco T289D mutant from Thermococcus kodakarensis | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fujihashi, M, Nishitani, Y, Kiriyama, T, Miki, K. | | Deposit date: | 2014-01-29 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mutation design of thermophilic Rubisco based on the three-dimensional structure enhances its activity at ambient temperature
to be published
|
|
5YSQ
 
 | | Sulfate-complex structure of a pyrophosphate-dependent kinase in the ribokinase family provides insight into the donor-binding mode | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, SULFATE ION, TM0415 | | Authors: | Nagata, R, Fujihashi, M, Miki, K. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Identification of a pyrophosphate-dependent kinase and its donor selectivity determinants.
Nat Commun, 9, 2018
|
|
5YSP
 
 | | Pyrophosphate-dependent kinase in the ribokinase family complexed with a pyrophosphate analog and myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, MAGNESIUM ION, METHYLENEDIPHOSPHONIC ACID, ... | | Authors: | Nagata, R, Fujihashi, M, Miki, K. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of a pyrophosphate-dependent kinase and its donor selectivity determinants.
Nat Commun, 9, 2018
|
|
