6Q21
 
 | |
6APM
 
 | | Hen egg-white lysozyme (WT), solved with serial millisecond crystallography using synchrotron radiation | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Lyubimov, A.Y, Mathews, I.I, Uervivojnangkoorn, M, Soltis, S.M, Cohen, A.E. | | Deposit date: | 2017-08-17 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Conformational Flexibility of the Acyltransferase from the Disorazole Polyketide Synthase Is Revealed by an X-ray Free-Electron Laser Using a Room-Temperature Sample Delivery Method for Serial Crystallography.
Biochemistry, 56, 2017
|
|
6APK
 
 | | Trans-acting transferase from Disorazole synthase solved by serial femtosecond XFEL crystallography | | Descriptor: | DisD protein | | Authors: | Lyubimov, A.Y, Mathews, I.I, Uervivojnangkoorn, M, Khosla, C, Soltis, S.M, Cohen, A.E. | | Deposit date: | 2017-08-17 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Conformational Flexibility of the Acyltransferase from the Disorazole Polyketide Synthase Is Revealed by an X-ray Free-Electron Laser Using a Room-Temperature Sample Delivery Method for Serial Crystallography.
Biochemistry, 56, 2017
|
|
6APF
 
 | | Trans-acting transferase from Disorazole synthase complexed with Citrate. | | Descriptor: | CITRIC ACID, DisD protein, GLYCEROL, ... | | Authors: | Mathews, I.I, Lyubimov, A, Soltis, M, Khosla, C, Cohen, A, Robbins, T. | | Deposit date: | 2017-08-17 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Conformational Flexibility of the Acyltransferase from the Disorazole Polyketide Synthase Is Revealed by an X-ray Free-Electron Laser Using a Room-Temperature Sample Delivery Method for Serial Crystallography.
Biochemistry, 56, 2017
|
|
6APG
 
 | | Trans-acting transferase from Disorazole synthase with malonate | | Descriptor: | CALCIUM ION, DisD protein, GLYCEROL, ... | | Authors: | Mathews, I.I, Lyubimov, A.Y, Soltis, S.M, Khosla, C, Robbins, T, Cohen, A.E. | | Deposit date: | 2017-08-17 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Conformational Flexibility of the Acyltransferase from the Disorazole Polyketide Synthase Is Revealed by an X-ray Free-Electron Laser Using a Room-Temperature Sample Delivery Method for Serial Crystallography.
Biochemistry, 56, 2017
|
|
5LP0
 
 | |
3CTI
 
 | |
3HMG
 
 | | REFINEMENT OF THE INFLUENZA VIRUS HEMAGGLUTININ BY SIMULATED ANNEALING | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Weis, W.I, Bruenger, A.T, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1989-09-11 | | Release date: | 1991-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Refinement of the influenza virus hemagglutinin by simulated annealing.
J.Mol.Biol., 212, 1990
|
|
3NF3
 
 | | Crystal structure of BoNT/A LC with JTH-NB-7239 peptide | | Descriptor: | BoNT/A, JTH-NB72-39 inhibitor, NICKEL (II) ION, ... | | Authors: | Zuniga, J.E. | | Deposit date: | 2010-06-09 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Iterative structure-based peptide-like inhibitor design against the botulinum neurotoxin serotype A.
Plos One, 5, 2010
|
|
3DS9
 
 | |
3DSE
 
 | |
1WTL
 
 | |
1WS1
 
 | |
1ELH
 
 | |
1ZXJ
 
 | | Crystal structure of the hypthetical Mycoplasma protein, MPN555 | | Descriptor: | Hypothetical protein MG377 homolog | | Authors: | Schulze-Gahmen, U, Aono, S, Shengfeng, C, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2005-06-08 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the hypothetical Mycoplasma protein MPN555 suggests a chaperone function.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1OWN
 
 | | DATA3:DNA photolyase / received X-rays dose 4.8 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWM
 
 | | DATA1:DNA photolyase / received X-rays dose 1.2 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWP
 
 | | DATA6:photoreduced DNA pholyase / received X-rays dose 4.8 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWL
 
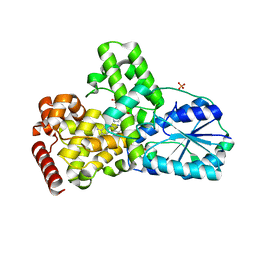 | | Structure of apophotolyase from Anacystis nidulans | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1N51
 
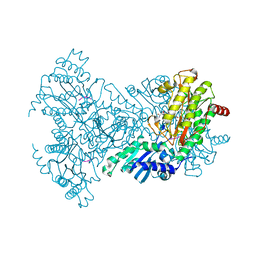 | | Aminopeptidase P in complex with the inhibitor apstatin | | Descriptor: | MANGANESE (II) ION, Xaa-Pro aminopeptidase, apstatin | | Authors: | Graham, S.C, Maher, M.J, Lee, M.H, Simmons, W.H, Freeman, H.C, Guss, J.M. | | Deposit date: | 2002-11-03 | | Release date: | 2003-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Escherichia coli aminopeptidase P in complex with the inhibitor apstatin.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OWO
 
 | | DATA4:photoreduced DNA photolyase / received X-rays dose 1.2 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1BFE
 
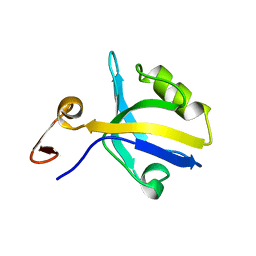 | | THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 | | Descriptor: | PSD-95 | | Authors: | Doyle, D.A, Lee, A, Lewis, J, Kim, E, Sheng, M, Mackinnon, R. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.
Cell(Cambridge,Mass.), 85, 1996
|
|
1BE9
 
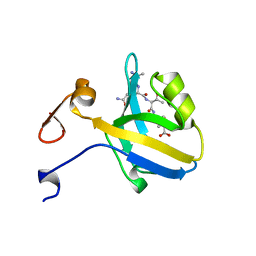 | | THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 IN COMPLEX WITH A C-TERMINAL PEPTIDE DERIVED FROM CRIPT. | | Descriptor: | CRIPT, PSD-95 | | Authors: | Doyle, D.A, Lee, A, Lewis, J, Kim, E, Sheng, M, Mackinnon, R. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.
Cell(Cambridge,Mass.), 85, 1996
|
|
1FL1
 
 | | KSHV PROTEASE | | Descriptor: | POTASSIUM ION, PROTEASE | | Authors: | Reiling, K.K, Pray, T.R, Craik, C.S, Stroud, R.M. | | Deposit date: | 2000-08-11 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional consequences of the Kaposi's sarcoma-associated herpesvirus protease structure: regulation of activity and dimerization by conserved structural elements.
Biochemistry, 39, 2000
|
|
1Q9E
 
 | | RNase T1 variant with adenine specificity | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Guanyl-specific ribonuclease T1 precursor | | Authors: | Czaja, R, Struhalla, M, Hoeschler, K, Saenger, W, Straeter, N, Hahn, U. | | Deposit date: | 2003-08-25 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RNase T1 Variant RV Cleaves Single-Stranded RNA after Purines Due to Specific Recognition by the Asn46 Side Chain Amide.
Biochemistry, 43, 2004
|
|
