7VKG
 
 | |
6L08
 
 | |
8EGQ
 
 | | Branched chain ketoacid dehydrogenase kinase complexes | | Descriptor: | (2S)-2-ethyl-4-{[(2'M)-2'-(1H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydro-2H-pyrido[3,2-b][1,4]oxazine, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
8EGF
 
 | | Branched chain ketoacid dehydrogenase kinase in complex with inhibitor | | Descriptor: | (5P)-5-(4'-methyl[1,1'-biphenyl]-2-yl)-1H-tetrazole, ADENOSINE-5'-DIPHOSPHATE, POTASSIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-12 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
8EGD
 
 | | Branched chain ketoacid dehydrogenase kinase in complex with inhibitor | | Descriptor: | 5-(4-methoxyphenyl)-1H-tetrazole, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-12 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
8EGU
 
 | | Branched chain ketoacid dehydrogenase kinase complexes | | Descriptor: | (2~{S})-3-methyl-2-[pentanoyl-[[4-[2-(2~{H}-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]methyl]amino]butanoic acid, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
5XAF
 
 | | Crystal structure of tubulin-stathmin-TTL-Compound Z1 complex | | Descriptor: | (3S,4R)-4-(3-hydroxy-4-methoxyphenyl)-3-methyl-1-(3,4,5-trimethoxyphenyl)azetidin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhang, H, Luo, C, Wang, Y. | | Deposit date: | 2017-03-12 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Design, synthesis, biological evaluation and cocrystal structures with tubulin of chiral beta-lactam bridged combretastatin A-4 analogues as potent antitumor agents
Eur J Med Chem, 144, 2017
|
|
4PP9
 
 | |
4MF0
 
 | | ITK kinase domain in complex with benzothiazole inhibitor compound 12a (1S,2S)-2-{4-[(DIMETHYLAMINO)METHYL]PHENYL}-N-[6-(PYRIDIN-3-YL)-1,3-BENZOTHIAZOL-2-YL]CYCLOPROPANECARBOXAMIDE (12a) | | Descriptor: | (1S,2S)-2-{4-[(dimethylamino)methyl]phenyl}-N-[6-(pyridin-3-yl)-1,3-benzothiazol-2-yl]cyclopropanecarboxamide, Tyrosine-protein kinase ITK/TSK | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2013-08-27 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structure-based design and synthesis of potent benzothiazole inhibitors of interleukin-2 inducible T cell kinase (ITK).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8F92
 
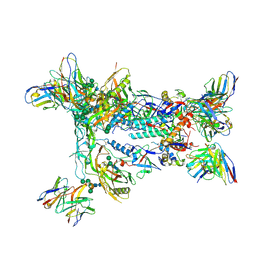 | | HIV Env BG505_MD39_B11 SOSIP boosting trimer in complex with B11_d77.7 mouse Fab and RM20A3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B11_d77.7 Fab heavy chain, ... | | Authors: | Torres, J.L, Ozorowski, G, Ward, A.B. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | mRNA-LNP HIV-1 trimer boosters elicit precursors to broad neutralizing antibodies.
Science, 384, 2024
|
|
8F9M
 
 | |
8F9G
 
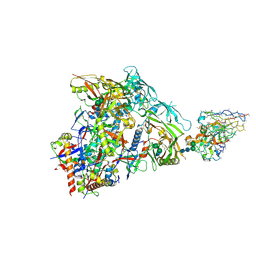 | | HIV Env germline targeting BG505_MD64_N332-GT5 SOSIP in complex with V3-glycan polyclonal Fab isolated from immunized BG18HCgl knock-in mice | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505_MD64_N332-GT5 gp120, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | mRNA-LNP HIV-1 trimer boosters elicit precursors to broad neutralizing antibodies.
Science, 384, 2024
|
|
2LWF
 
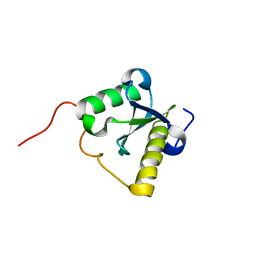 | | Structure of N-terminal domain of a plant Grx | | Descriptor: | Monothiol glutaredoxin-S16, chloroplastic | | Authors: | Feng, Y. | | Deposit date: | 2012-07-28 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the N-terminal GIY-YIG endonuclease activity of Arabidopsis glutaredoxin AtGRXS16 in chloroplasts.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4L1B
 
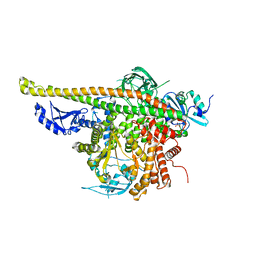 | | Crystal Structure of p110alpha complexed with niSH2 of p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, SULFATE ION | | Authors: | Zhang, J, Zhao, Y.L, Chen, Y.Y, Huang, M, Jiang, F. | | Deposit date: | 2013-06-03 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Crystal Structures of PI3K alpha Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design.
ACS Med Chem Lett, 5, 2014
|
|
7DCN
 
 | | Apo-citrate lyase phosphoribosyl-dephospho-CoA transferase | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, SULFATE ION, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
7DCM
 
 | | Crystal structure of CITX | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
4L2Y
 
 | | Crystal Structure of p110alpha complexed with niSH2 of p85alpha and compound 9d | | Descriptor: | 3-amino-5-[4-(morpholin-4-yl)pyrido[3',2':4,5]furo[3,2-d]pyrimidin-2-yl]phenol, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Zhang, J, Zhao, Y.L, Chen, Y.Y, Huang, M, Jiang, F. | | Deposit date: | 2013-06-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of PI3K alpha Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design.
ACS Med Chem Lett, 5, 2014
|
|
4RFM
 
 | | ITK kinase domain in complex with compound 1 N-{1-[(1,1-dioxo-1-thian-2-yl)(phenyl)methyl]-1H- pyrazol-4-yl}-5,5-difluoro-5a-methyl-1H,4H,4aH,5H,5aH,6H-cyclopropa[f]indazole-3-carboxamide | | Descriptor: | (4aS,5aR)-N-{1-[(R)-[(2R)-1,1-dioxidotetrahydro-2H-thiopyran-2-yl](phenyl)methyl]-1H-pyrazol-4-yl}-5,5-difluoro-5a-methyl-1,4,4a,5,5a,6-hexahydrocyclopropa[f]indazole-3-carboxamide, Tyrosine-protein kinase ITK/TSK | | Authors: | McEwan, P.A, Barker, J.J, Eigenbrot, C. | | Deposit date: | 2014-09-26 | | Release date: | 2015-04-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tetrahydroindazoles as Interleukin-2 Inducible T-Cell Kinase Inhibitors. Part II. Second-Generation Analogues with Enhanced Potency, Selectivity, and Pharmacodynamic Modulation in Vivo.
J.Med.Chem., 58, 2015
|
|
8HSB
 
 | |
7CZP
 
 | | S protein of SARS-CoV-2 in complex bound with P2B-1A1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c181_light_IGLV2-14_IGLJ3,IGL@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZQ
 
 | | S protein of SARS-CoV-2 in complex bound with P2B-1A10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c642_heavy_IGHV3-53_IGHD1-26_IGHJ6,Chain H of P2B-1A10,Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZR
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-1B8_2B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c542_heavy_IGHV3-53_IGHD3-10_IGHJ6,IGH@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZT
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-2G9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c689_light_IGLV5-37_IGLJ3,IGL@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZS
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-1B8_3B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c542_heavy_IGHV3-53_IGHD3-10_IGHJ6,IGH@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZU
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-1B6_2B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy variable 3-30-3,Immunoglobulin heavy variable 3-30-3,Chain H of P5A-1B6_2B,Immunoglobulin gamma-1 heavy chain,Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
