4CNN
 
 | | High resolution structure of Salmonella typhi type I dehydroquinase | | Descriptor: | 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Lence, E, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-01-23 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Mechanistic Insight Into the Reaction Catalyzed by Type I Dehydroquinase
To be Published
|
|
6A2T
 
 | | Mycobacterium tuberculosis LexA C-domain K197A | | Descriptor: | ACRYLIC ACID, LexA repressor | | Authors: | Chandran, A.V, Srikalaivani, R, Paul, A, Vijayan, M. | | Deposit date: | 2018-06-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical characterization of Mycobacterium tuberculosis LexA and structural studies of its C-terminal segment.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6A6N
 
 | | Crystal structure of an inward-open apo state of the eukaryotic ABC multidrug transporter CmABCB1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP-binding cassette, sub-family B, ... | | Authors: | Kato, H, Nakatsu, T, Kodan, A. | | Deposit date: | 2018-06-28 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Inward- and outward-facing X-ray crystal structures of homodimeric P-glycoprotein CmABCB1.
Nat Commun, 10, 2019
|
|
4CSH
 
 | | Native structure of the lytic CHAPK domain of the endolysin LysK from Staphylococcus aureus bacteriophage K | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Sanz-Gaitero, M, Keary, R, Garcia-Doval, C, Coffey, A, van Raaij, M.J. | | Deposit date: | 2014-03-07 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the lytic CHAP(K) domain of the endolysin LysK from Staphylococcus aureus bacteriophage K.
Virol. J., 11, 2014
|
|
6A75
 
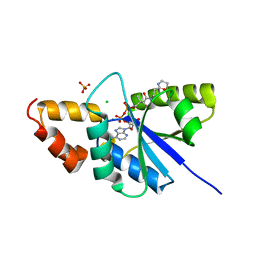 | | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephospho Coenzyme A at 2.75 A resolution | | Descriptor: | CHLORIDE ION, DEPHOSPHO COENZYME A, MAGNESIUM ION, ... | | Authors: | Singh, P.K, Gupta, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2018-07-02 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephospho Coenzyme A at 2.75 A resolution
To Be Published
|
|
6A7F
 
 | |
6A9U
 
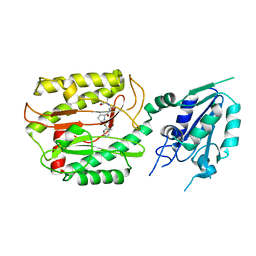 | | Crystal strcture of Icp55 from Saccharomyces cerevisiae bound to apstatin inhibitor | | Descriptor: | Intermediate cleaving peptidase 55, MANGANESE (II) ION, apstatin | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
6A89
 
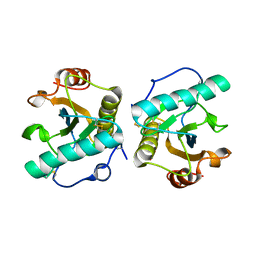 | | Crystal structure of the ternary complex of peptidoglycan recognition protein (PGRP-S) with Tartaric acid, Ribose and 2,6-DIAMINOPIMELIC ACID at 2.11 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2,6-DIAMINOPIMELIC ACID, GLYCEROL, ... | | Authors: | Bairagya, H.R, Shokeen, A, Sharma, P, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2018-07-06 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of the ternary complex of peptidoglycan recognition protein (PGRP-S) with Tartaric acid, Ribose and 2,6-DIAMINOPIMELIC ACID at 2.11 A resolution
To Be Published
|
|
1S4Z
 
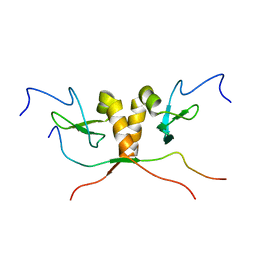 | | HP1 chromo shadow domain in complex with PXVXL motif of CAF-1 | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromobox protein homolog 1 | | Authors: | Thiru, A, Nietlispach, D, Mott, H.R, Okuwaki, M, Lyon, D, Nielsen, P.R, Hirshberg, M, Verreault, A, Murzina, N.V, Laue, E.D. | | Deposit date: | 2004-01-19 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin.
Embo J., 23, 2004
|
|
4CKR
 
 | | Crystal structure of the human DDR1 kinase domain in complex with DDR1-IN-1 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-ethylpiperazin-1-yl)methyl]-n-{4-methyl-3-[(2-oxo-2,3-dihydro-1h-indol-5-yl)oxy]phenyl}-3-(trifluoromethyl)benzamide, EPITHELIAL DISCOIDIN DOMAIN-CONTAINING RECEPTOR 1 | | Authors: | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Krojer, T, Newman, J.A, Dixon-Clarke, S, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Potent and Selective Ddr1 Receptor Tyrosine Kinase Inhibitor.
Acs Chem.Biol., 8, 2013
|
|
6A9V
 
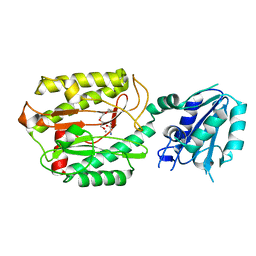 | | Crystal structure of Icp55 from Saccharomyces cerevisiae (N-terminal 42 residues deletion) | | Descriptor: | GLYCINE, Intermediate cleaving peptidase 55, MANGANESE (II) ION, ... | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
1S9I
 
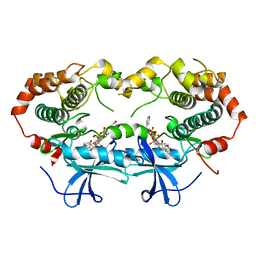 | | X-ray structure of the human mitogen-activated protein kinase kinase 2 (MEK2)in a complex with ligand and MgATP | | Descriptor: | 5-{3,4-DIFLUORO-2-[(2-FLUORO-4-IODOPHENYL)AMINO]PHENYL}-N-(2-MORPHOLIN-4-YLETHYL)-1,3,4-OXADIAZOL-2-AMINE, ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 2, ... | | Authors: | Ohren, J.F, Chen, H, Pavlovsky, A, Whitehead, C, Yan, C, McConnell, P, Delaney, A, Dudley, D.T, Sebolt-Leopold, J, Hasemann, C.A. | | Deposit date: | 2004-02-04 | | Release date: | 2004-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition.
Nat.Struct.Mol.Biol., 11, 2004
|
|
6ABZ
 
 | | Crystal Structure of HEWL in deionized water | | Descriptor: | CHLORIDE ION, Lysozyme C, S-1,2-PROPANEDIOL, ... | | Authors: | Seyedarabi, A, Seraj, Z. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The role of Cinnamaldehyde and Phenyl ethyl alcohol as two types of precipitants affecting protein hydration levels.
Int.J.Biol.Macromol., 146, 2019
|
|
4CLM
 
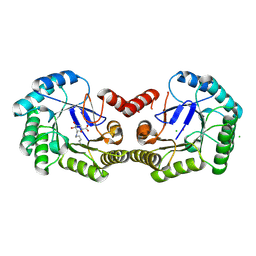 | | Structure of Salmonella typhi type I dehydroquinase irreversibly inhibited with a 1,3,4-trihydroxyciclohexane-1-carboxylic acid derivative | | Descriptor: | (1~{S},3~{S},4~{R},5~{R})-3-methyl-1,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Tizon, L, Maneiro, M, Lence, E, Poza, S, Lamb, H, Hawkins, A.R, Blanco, B, Sedes, A, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-01-15 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Irreversible covalent modification of type I dehydroquinase with a stable Schiff base.
Org. Biomol. Chem., 13, 2015
|
|
6AGR
 
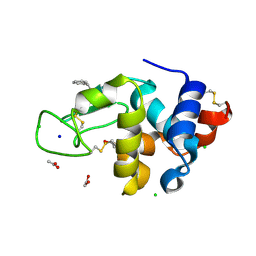 | | Structure of HEWL co-crystallised with phenylethyl alcohol | | Descriptor: | 1,2-ETHANEDIOL, 2-PHENYL-ETHANOL, ACETATE ION, ... | | Authors: | Seyedarabi, A, Seraj, Z. | | Deposit date: | 2018-08-13 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | The role of Cinnamaldehyde and Phenyl ethyl alcohol as two types of precipitants affecting protein hydration levels.
Int.J.Biol.Macromol., 146, 2019
|
|
5ZRO
 
 | | M. smegmatis antimutator protein MutT2 in complex with 5mdCTP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXY-5-METHYLCYTIDINE 5'-(TETRAHYDROGEN TRIPHOSPHATE), Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
5ZWE
 
 | | Covalent bond formation between histidine of Vitamin D receptor (VDR) and a full agonist having a vinyl ketone group via conjugate addition reaction | | Descriptor: | (6R)-6-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]hept-1-en-3-one, 13-meric peptide from DRIP205 NR2 BOX peptide, Vitamin D3 receptor | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
6AIF
 
 | |
5ZYB
 
 | |
6AL5
 
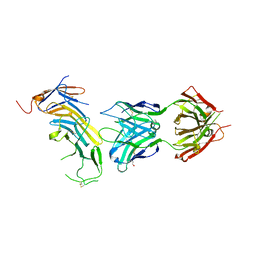 | | COMPLEX BETWEEN CD19 (N138Q MUTANT) AND B43 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, B-lymphocyte antigen CD19, B43 HEAVY CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of B-cell co-receptor CD19 in complex with antibody B43 reveals an unexpected fold.
Proteins, 86, 2018
|
|
6A1Q
 
 | | Crystal structures of disordered Z-type helices | | Descriptor: | DNA (5'-D(P*CP*AP*CP*A)-3'), DNA (5'-D(P*TP*GP*TP*G)-3') | | Authors: | Karthik, S, Mandal, P.K, Thirugnanasambandam, A, Gautham, N. | | Deposit date: | 2018-06-07 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal structures of disordered Z-type helices.
Nucleosides Nucleotides Nucleic Acids, 38, 2019
|
|
6A8I
 
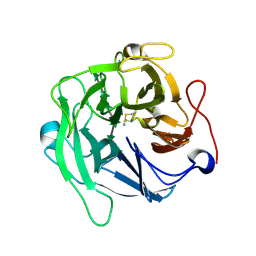 | | Crystal structure of endo-arabinanase ABN-TS D147N mutant in complex with arabinohexaose | | Descriptor: | CALCIUM ION, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose, endo-alpha-(1->5)-L-arabinanase | | Authors: | Yamaguchi, A, Tada, T. | | Deposit date: | 2018-07-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of endo-1,5-alpha-L-arabinanase mutants from Bacillus thermodenitrificans TS-3 in complex with arabino-oligosaccharides.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6A8U
 
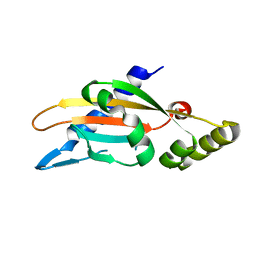 | | PhoQ sensor domain (wild type): analysis of internal cavity | | Descriptor: | Sensor protein PhoQ | | Authors: | Yoshitani, K, Ishii, E, Taniguchi, K, Sugimoto, H, Shiro, Y, Mori, H, Akiyama, Y, Kato, A, Utsumi, R, Eguchi, Y. | | Deposit date: | 2018-07-10 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Identification of an internal cavity in the PhoQ sensor domain for PhoQ activity and SafA-mediated control.
Biosci. Biotechnol. Biochem., 83, 2019
|
|
4BNI
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 2-(2- aminophenoxy)-5-hexylphenol | | Descriptor: | 2-(2-azanylphenoxy)-5-hexyl-phenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
2JDU
 
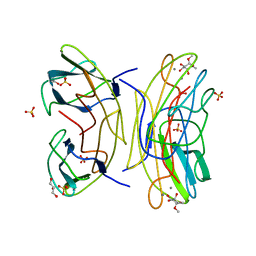 | | Mutant (G24N) of Pseudomonas aeruginosa lectin II (PA-IIL) complexed with methyl-a-L-fucopyranoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, GLYCEROL, ... | | Authors: | Adam, J, Pokorna, M, Sabin, C, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2007-01-12 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering of Pa-Iil Lectin from Pseudomonas Aeruginosa - Unravelling the Role of the Specificity Loop for Sugar Preference.
Bmc Struct.Biol., 7, 2007
|
|
