2WVT
 
 | | Crystal structure of an alpha-L-fucosidase GH29 from Bacteroides thetaiotaomicron in complex with a novel iminosugar fucosidase inhibitor | | Descriptor: | (2S,3R,5R,6S)-3,4,5-TRIHYDROXY-2,6-BIS(HYDROXYMETHYL)PIPERIDINIUM, ALPHA-L-FUCOSIDASE, GLYCEROL, ... | | Authors: | Lammerts van Bueren, A, Ardevol, A, Fayers-Kerr, J, Luo, B, Zhang, Y, Sollogoub, M, Bleriot, Y, Rovira, C, Davies, G.J. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of the Reaction Coordinate of Alpha-L-Fucosidases: A Combined Structural and Quantum Mechanical Approach.
J.Am.Chem.Soc., 132, 2010
|
|
3TUD
 
 | | Crystal structure of SYK kinase domain with N-(4-methyl-3-(8-methyl-7-oxo-2-(phenylamino)-7,8-dihydropyrido[2,3-d]pyrimidin-6-yl)phenyl)-3-(trifluoromethyl)benzamide | | Descriptor: | N-{4-methyl-3-[8-methyl-7-oxo-2-(phenylamino)-7,8-dihydropyrido[2,3-d]pyrimidin-6-yl]phenyl}-3-(trifluoromethyl)benzamide, Tyrosine-protein kinase SYK | | Authors: | Lovering, F, McDonald, J, Whitlock, G, Glossop, P, Phillips, C, Sabnis, Y, Ryan, M, Fitz, L, Lee, J, Chang, J.S, Han, S, Kurumbail, R, Thorarenson, A. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Identification of Type-II Inhibitors Using Kinase Structures.
Chem.Biol.Drug Des., 80, 2012
|
|
3TUC
 
 | | Crystal structure of SYK kinase domain with 1-benzyl-N-(5-(6,7-dimethoxyquinolin-4-yloxy)pyridin-2-yl)-2-oxo-1,2-dihydropyridine-3-carboxamide | | Descriptor: | 1-benzyl-N-{5-[(6,7-dimethoxyquinolin-4-yl)oxy]pyridin-2-yl}-2-oxo-1,2-dihydropyridine-3-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Lovering, F, McDonald, J, Whitlock, G, Glossop, P, Phillips, C, Sabnis, Y, Ryan, M, Fitz, L, Lee, J, Chang, J.S, Han, S, Kurumbail, R, Thorarenson, A. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Type-II Inhibitors Using Kinase Structures.
Chem.Biol.Drug Des., 80, 2012
|
|
1PLK
 
 | | CRYSTALLOGRAPHIC STUDIES ON P21H-RAS USING SYNCHROTRON LAUE METHOD: IMPROVEMENT OF CRYSTAL QUALITY AND MONITORING OF THE GTPASE REACTION AT DIFFERENT TIME POINTS | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Scheidig, A, Sanchez-Llorente, A, Lautwein, A, Pai, E.F, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Goody, R.S. | | Deposit date: | 1994-03-13 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic studies on p21(H-ras) using the synchrotron Laue method: improvement of crystal quality and monitoring of the GTPase reaction at different time points.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1PLJ
 
 | | CRYSTALLOGRAPHIC STUDIES ON P21H-RAS USING SYNCHROTRON LAUE METHOD: IMPROVEMENT OF CRYSTAL QUALITY AND MONITORING OF THE GTPASE REACTION AT DIFFERENT TIME POINTS | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE 5'-TRIPHOSPHATE P3-[1-(2-NITROPHENYL)ETHYL ESTER], MAGNESIUM ION | | Authors: | Scheidig, A, Sanchez-Llorente, A, Lautwein, A, Pai, E.F, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Goody, R.S. | | Deposit date: | 1994-03-13 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic studies on p21(H-ras) using the synchrotron Laue method: improvement of crystal quality and monitoring of the GTPase reaction at different time points.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1PLL
 
 | | CRYSTALLOGRAPHIC STUDIES ON P21H-RAS USING SYNCHROTRON LAUE METHOD: IMPROVEMENT OF CRYSTAL QUALITY AND MONITORING OF THE GTPASE REACTION AT DIFFERENT TIME POINTS | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Scheidig, A, Sanchez-Llorente, A, Lautwein, A, Pai, E.F, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Goody, R.S. | | Deposit date: | 1994-03-13 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic studies on p21(H-ras) using the synchrotron Laue method: improvement of crystal quality and monitoring of the GTPase reaction at different time points.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
3ZM8
 
 | | Crystal structure of Podospora anserina GH26-CBM35 beta-(1,4)- mannanase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GH26 ENDO-BETA-1,4-MANNANASE, ... | | Authors: | Couturier, M, Roussel, A, Rosengren, A, Leone, P, Stalbrand, H, Berrin, J.G. | | Deposit date: | 2013-02-06 | | Release date: | 2013-04-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and Biochemical Analyses of Glycoside Hydrolase Families 5 and 26 Beta-(1,4)-Mannanases from Podospora Anserina Reveal Differences Upon Manno-Oligosaccharides Catalysis.
J.Biol.Chem., 288, 2013
|
|
1EGO
 
 | | NMR STRUCTURE OF OXIDIZED ESCHERICHIA COLI GLUTAREDOXIN: COMPARISON WITH REDUCED E. COLI GLUTAREDOXIN AND FUNCTIONALLY RELATED PROTEINS | | Descriptor: | GLUTAREDOXIN | | Authors: | Xia, T.-H, Bushweller, J.H, Sodano, P, Billeter, M, Bjornberg, O, Holmgren, A, Wuthrich, K. | | Deposit date: | 1991-10-08 | | Release date: | 1993-10-31 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of oxidized Escherichia coli glutaredoxin: comparison with reduced E. coli glutaredoxin and functionally related proteins.
Protein Sci., 1, 1992
|
|
2QTJ
 
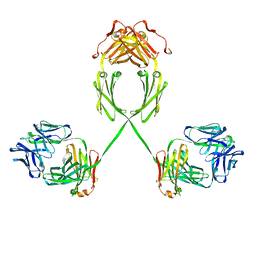 | | Solution structure of human dimeric immunoglobulin A | | Descriptor: | Ig alpha-1 chain C region, Kappa light chain IgA1 | | Authors: | Bonner, A, Furtado, P.B, Almogren, A, Kerr, M.A, Perkins, S.J. | | Deposit date: | 2007-08-02 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Implications of the near-planar solution structure of human myeloma dimeric IgA1 for mucosal immunity and IgA nephropathy
J.Immunol., 180, 2008
|
|
1EGR
 
 | | SEQUENCE-SPECIFIC 1H N.M.R. ASSIGNMENTS AND DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF REDUCED ESCHERICHIA COLI GLUTAREDOXIN | | Descriptor: | GLUTAREDOXIN | | Authors: | Sodano, P, Xia, T.-H, Bushweller, J.H, Bjornberg, O, Holmgren, A, Billeter, M, Wuthrich, K. | | Deposit date: | 1991-10-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific 1H n.m.r. assignments and determination of the three-dimensional structure of reduced Escherichia coli glutaredoxin.
J.Mol.Biol., 221, 1991
|
|
2RD3
 
 | |
2VZV
 
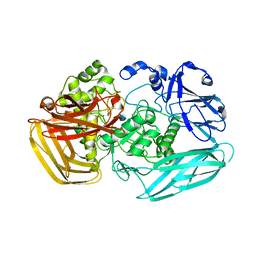 | | Substrate Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with chitosan | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, EXO-BETA-D-GLUCOSAMINIDASE | | Authors: | Lammerts van Bueren, A, Ghinet, M.G, Gregg, K, Fleury, A, Brzezinski, R, Boraston, A.B. | | Deposit date: | 2008-08-05 | | Release date: | 2008-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structural Basis of Substrate Recognition in an Exo-Beta-D-Glucosaminidase Involved in Chitosan Hydrolysis.
J.Mol.Biol., 385, 2009
|
|
2VZO
 
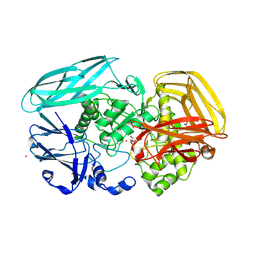 | | Crystal structure of Amycolatopsis orientalis exo-chitosanase CsxA | | Descriptor: | ACETATE ION, CADMIUM ION, EXO-BETA-D-GLUCOSAMINIDASE, ... | | Authors: | Lammerts van Bueren, A, Ghinet, M.G, Gregg, K, Fleury, A, Brzezinski, R, Boraston, A.B. | | Deposit date: | 2008-08-05 | | Release date: | 2008-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Structural Basis of Substrate Recognition in an Exo-Beta-D-Glucosaminidase Involved in Chitosan Hydrolysis.
J.Mol.Biol., 385, 2009
|
|
3EZR
 
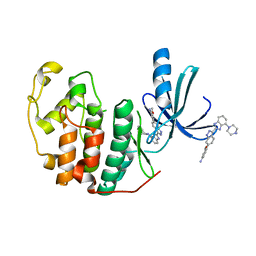 | | CDK-2 with indazole inhibitor 17 bound at its active site | | Descriptor: | 3-methoxy-4-{3-[4-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2 | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-10-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2VZU
 
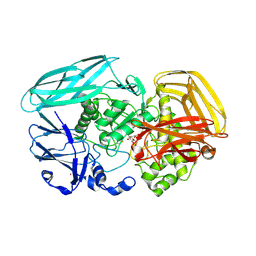 | | Complex of Amycolatopsis orientalis exo-chitosanase CsxA D469A with PNP-beta-D-glucosamine | | Descriptor: | 4-nitrophenyl 2-amino-2-deoxy-beta-D-glucopyranoside, ACETATE ION, CADMIUM ION, ... | | Authors: | Lammerts van Bueren, A, Ghinet, M.G, Gregg, K, Fleury, A, Brzezinski, R, Boraston, A.B. | | Deposit date: | 2008-08-05 | | Release date: | 2008-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structural Basis of Substrate Recognition in an Exo-Beta-D-Glucosaminidase Involved in Chitosan Hydrolysis.
J.Mol.Biol., 385, 2009
|
|
2VZS
 
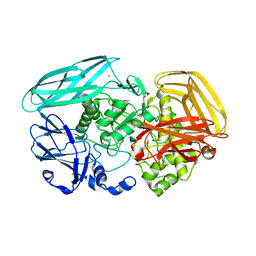 | | Chitosan Product complex of Amycolatopsis orientalis exo-chitosanase CsxA | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, CADMIUM ION, EXO-BETA-D-GLUCOSAMINIDASE, ... | | Authors: | Lammerts van Bueren, A, Ghinet, M.G, Gregg, K, Fleury, A, Brzezinski, R, Boraston, A.B. | | Deposit date: | 2008-08-05 | | Release date: | 2008-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structural Basis of Substrate Recognition in an Exo-Beta-D-Glucosaminidase Involved in Chitosan Hydrolysis.
J.Mol.Biol., 385, 2009
|
|
3F5X
 
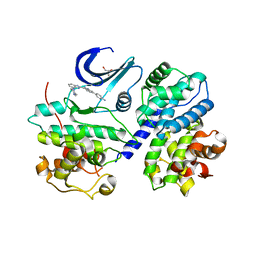 | | CDK-2-Cyclin complex with indazole inhibitor 9 bound at its active site | | Descriptor: | 4-{3-[7-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-11-04 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3EZV
 
 | | CDK-2 with indazole inhibitor 9 bound at its active site | | Descriptor: | 4-{3-[7-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2 | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-10-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3E1Z
 
 | | Crystal structure of the parasite protesase inhibitor chagasin in complex with papain | | Descriptor: | ACETIC ACID, Chagasin, FORMIC ACID, ... | | Authors: | Redzynia, I, Bujacz, G, Bujacz, A, Ljunggren, A, Abrahamson, M, Jaskolski, M. | | Deposit date: | 2008-08-05 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the parasite inhibitor chagasin in complex with papain allows identification of structural requirements for broad reactivity and specificity determinants for target proteases.
Febs J., 276, 2009
|
|
2WVV
 
 | | Crystal structure of an alpha-L-fucosidase GH29 from Bacteroides thetaiotaomicron | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-L-FUCOSIDASE, GLYCEROL, ... | | Authors: | Lammerts van Bueren, A, Ardevol, A, Fayers-Kerr, J, Luo, B, Zhang, Y, Sollogoub, M, Bleriot, Y, Rovira, C, Davies, G.J. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Analysis of the Reaction Coordinate of Alpha-L-Fucosidases: A Combined Structural and Quantum Mechanical Approach
J.Am.Chem.Soc., 132, 2010
|
|
2WVS
 
 | | Crystal structure of an alpha-L-fucosidase GH29 trapped covalent intermediate from Bacteroides thetaiotaomicron in complex with 2- fluoro-fucosyl fluoride using an E288Q mutant | | Descriptor: | 2-deoxy-2-fluoro-beta-L-fucopyranose, ALPHA-L-FUCOSIDASE, SULFATE ION | | Authors: | Lammerts van Bueren, A, Ardevol, A, Fayers-Kerr, J, Luo, B, Zhang, Y, Sollogoub, M, Bleriot, Y, Rovira, C, Davies, G.J. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Analysis of the Reaction Coordinate of Alpha-L-Fucosidases: A Combined Structural and Quantum Mechanical Approach
J.Am.Chem.Soc., 132, 2010
|
|
1NXD
 
 | | Crystal structure of MnMn Concanavalin A | | Descriptor: | AZIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lopez-Jaramillo, F.J, Gonzalez-Ramirez, L.A, Albert, A, Santoyo-Gonzalez, F, Vargas-Berenguel, A, Otalora, F. | | Deposit date: | 2003-02-10 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of concanavalin A at pH 8: bound solvent and crystal contacts.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2VZT
 
 | | Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with PNP-beta-D-glucosamine | | Descriptor: | 4-nitrophenyl 2-amino-2-deoxy-beta-D-glucopyranoside, ACETATE ION, CADMIUM ION, ... | | Authors: | Lammerts van Bueren, A, Ghinet, M.G, Gregg, K, Fleury, A, Brzezinski, R, Boraston, A.B. | | Deposit date: | 2008-08-05 | | Release date: | 2008-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis of Substrate Recognition in an Exo-Beta-D-Glucosaminidase Involved in Chitosan Hydrolysis.
J.Mol.Biol., 385, 2009
|
|
2WVU
 
 | | Crystal structure of a Michaelis complex of alpha-L-fucosidase GH29 from Bacteroides thetaiotaomicron with the synthetic substrate 4- nitrophenyl-alpha-L-fucose | | Descriptor: | 4-nitrophenyl 6-deoxy-alpha-L-galactopyranoside, ALPHA-L-FUCOSIDASE, SULFATE ION | | Authors: | Lammerts van Bueren, A, Ardevol, A, Fayers-Kerr, J, Luo, B, Zhang, Y, Sollogoub, M, Bleriot, Y, Rovira, C, Davies, G.J. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of the Reaction Coordinate of Alpha-L-Fucosidases: A Combined Structural and Quantum Mechanical Approach
J.Am.Chem.Soc., 132, 2010
|
|
2YA2
 
 | | Catalytic Module of the Multi-modular glycogen-degrading pneumococcal virulence factor SpuA in complex with an inhibitor. | | Descriptor: | CALCIUM ION, PUTATIVE ALKALINE AMYLOPULLULANASE, SODIUM ION, ... | | Authors: | Lammerts van Bueren, A, Ficko-Blean, E, Pluvinage, B, Hehemann, J.H, Higgins, M.A, Deng, L, Ogunniyi, A.D, Stroeher, U.H, Warry, N.E, Burke, R.D, Czjzek, M, Paton, J.C, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The Conformation and Function of a Multimodular Glycogen-Degrading Pneumococcal Virulence Factor.
Structure, 19, 2011
|
|
