1QN8
 
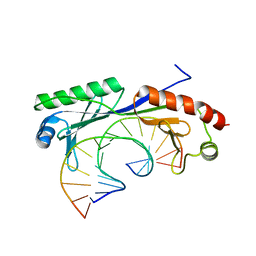 | | Crystal structure of the T(-28) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*TP*AP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*TP*TP*AP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | Authors: | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1QN5
 
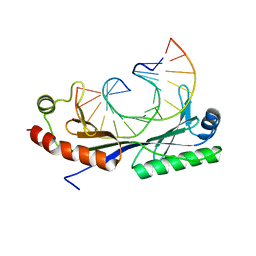 | | Crystal structure of the G(-26) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*GP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*CP*TP*TP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | Authors: | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | Deposit date: | 1999-10-14 | | Release date: | 2000-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
1OMI
 
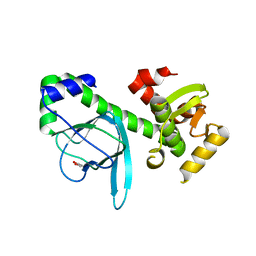 | | Crystal structure of PrfA,the transcriptional regulator in Listeria monocytogenes | | Descriptor: | GLYCEROL, Listeriolysin regulatory protein | | Authors: | Thirumuruhan, R, Rajashankar, K, Fedorov, A.A, Dodatko, T, Chance, M.R, Cossart, P, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-02-25 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of PrfA, the transcriptional regulator in Listeria monocytogenes
To be Published
|
|
5Z9W
 
 | | Ebola virus nucleoprotein-RNA complex | | Descriptor: | Ebolavirus nucleoprotein (residues 19-406), RNA (6-MER) | | Authors: | Sugita, Y, Matsunami, H, Kawaoka, Y, Noda, T, Wolf, M. | | Deposit date: | 2018-02-05 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the Ebola virus nucleoprotein-RNA complex at 3.6 angstrom resolution.
Nature, 563, 2018
|
|
6AVK
 
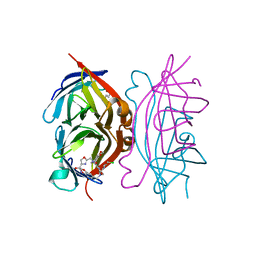 | | Streptavidin bound to peptide-like compound KPM-6 | | Descriptor: | 1-hydroxydodecan-4-one, N-[(2H-1,3-benzodioxol-5-yl)methyl]-2-({[(2H-1,3-benzodioxol-5-yl)methyl][2-(chloromethyl)-1,3-oxazole-4-carbonyl]amino}methyl)-N-[(4-carbamoyl-1,3-oxazol-2-yl)methyl]-1,3-oxazole-4-carboxamide, Streptavidin | | Authors: | Park, H, Shamim, R, McEnaney, P, Kodadek, T. | | Deposit date: | 2017-09-02 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Efficient Workflow for Screening DNA-encoded One Bead One Compound Libraries Using a Flow Cytometer
To be Published
|
|
7Z20
 
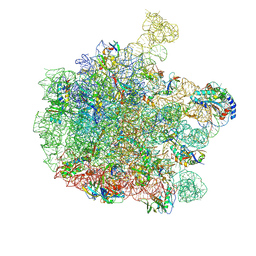 | | 70S E. coli ribosome with an extended uL23 loop from Candidatus marinimicrobia and a stalled filamin domain 5 nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Plessa, E, Wlodarski, T, Ahn, M, Sidhu, H, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-02-25 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
1V94
 
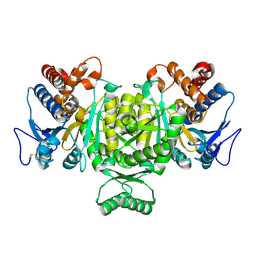 | | Crystal structure of isocitrate dehydrogenase from Aeropyrum pernix | | Descriptor: | isocitrate dehydrogenase | | Authors: | Jeong, J.-J, Sonoda, T, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2004-01-20 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of isocitrate dehydrogenase from Aeropyrum pernix
Proteins, 55, 2004
|
|
2VUL
 
 | | Thermostable mutant of ENVIRONMENTALLY ISOLATED GH11 XYLANASE | | Descriptor: | DODECAETHYLENE GLYCOL, GH11 XYLANASE, SULFATE ION | | Authors: | Dumon, C, Varvak, A, Wall, M.A, Flint, J.E, Lewis, R.J, Lakey, J.H, Luginbuhl, P, Healey, S, Todaro, T, Desantis, G, Sun, M, Parra-Gessert, L, Tan, X, Weiner, D.P, Gilbert, H.J. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering Hyperthermostability Into a Gh11 Xylanase is Mediated by Subtle Changes to Protein Structure.
J.Biol.Chem., 283, 2008
|
|
2ZW0
 
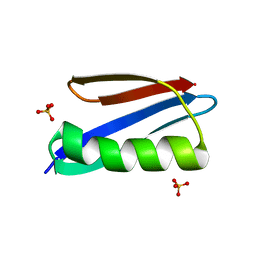 | | Crystal structure of a Streptococcal protein G B1 mutant | | Descriptor: | Protein LG, SULFATE ION | | Authors: | Watanabe, H, Matsumaru, H, Odahara, T, Suto, K, Honda, S. | | Deposit date: | 2008-11-26 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Optimizing pH response of affinity between protein G and IgG Fc: how electrostatic modulations affect protein-protein interactions.
J.Biol.Chem., 284, 2009
|
|
2ZW1
 
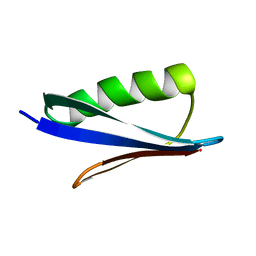 | | Crystal structure of a Streptococcal protein G B1 mutant | | Descriptor: | Protein LG | | Authors: | Watanabe, H, Matsumaru, H, Odahara, T, Suto, K, Honda, S. | | Deposit date: | 2008-11-26 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Optimizing pH response of affinity between protein G and IgG Fc: how electrostatic modulations affect protein-protein interactions.
J.Biol.Chem., 284, 2009
|
|
1WUA
 
 | | The structure of Aplyronine A-actin complex | | Descriptor: | (8R,9R,10R,11R,14S,18S,20S,24S)-24-{(1R,2S,3R,6R,7R,8R,9S,10E)-8-(ACETYLOXY)-6-[(N,N-DIMETHYLALANYL)OXY]-11-[FORMYL(MET HYL)AMINO]-2-HYDROXY-1,3,7,9-TETRAMETHYLUNDEC-10-ENYL}-10-HYDROXY-14,20-DIMETHOXY-9,11,15,18-TETRAMETHYL-2-OXOOXACYCLOTE TRACOSA-3,5,15,21-TETRAEN-8-YL N,N,O-TRIMETHYLSERINATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Hirata, K, Muraoka, S, Suenaga, K, Kuroda, T, Kato, K, Tanaka, H, Yamamoto, M, Takata, M, Yamada, K, Kigoshi, H. | | Deposit date: | 2004-12-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure basis for antitumor effect of aplyronine a
J.Mol.Biol., 356, 2006
|
|
1K8F
 
 | | CRYSTAL STRUCTURE OF THE HUMAN C-TERMINAL CAP1-ADENYLYL CYCLASE ASSOCIATED PROTEIN | | Descriptor: | ADENYLYL CYCLASE-ASSOCIATED PROTEIN | | Authors: | Patskovsky, Y.V, Chance, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-10-24 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the actin binding domain of the cyclase-associated protein.
Biochemistry, 43, 2004
|
|
5B0V
 
 | | Crystal Structure of Marburg virus VP40 Dimer | | Descriptor: | ETHANOL, Matrix protein VP40 | | Authors: | Oda, S, Bornholdt, Z.A, Abelson, D.M, Saphire, E.O. | | Deposit date: | 2015-11-05 | | Release date: | 2016-01-20 | | Last modified: | 2016-03-09 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal Structure of Marburg Virus VP40 Reveals a Broad, Basic Patch for Matrix Assembly and a Requirement of the N-Terminal Domain for Immunosuppression
J.Virol., 90, 2015
|
|
6LUH
 
 | | High resolution structure of N(omega)-hydroxy-L-arginine hydrolase | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, N(omega)-hydroxy-L-arginine amidinohydrolase | | Authors: | Oda, K, Matoba, Y. | | Deposit date: | 2020-01-28 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an Nomega-hydroxy-L-arginine hydrolase found in the D-cycloserine biosynthetic pathway.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6LUG
 
 | | Crystal structure of N(omega)-hydroxy-L-arginine hydrolase | | Descriptor: | MANGANESE (II) ION, N(omega)-hydroxy-L-arginine amidinohydrolase | | Authors: | Oda, K, Matoba, Y. | | Deposit date: | 2020-01-28 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an Nomega-hydroxy-L-arginine hydrolase found in the D-cycloserine biosynthetic pathway.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
3AN2
 
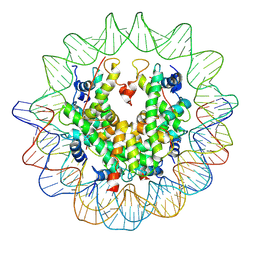 | | The structure of the centromeric nucleosome containing CENP-A | | Descriptor: | 147 mer DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Tachiwana, H, Kagawa, W, Shiga, T, Saito, K, Osakabe, A, Hayashi-Takanaka, Y, Park, S.-Y, Kimura, H, Kurumizaka, H. | | Deposit date: | 2010-08-27 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of the human centromeric nucleosome containing CENP-A
Nature, 476, 2011
|
|
3PDM
 
 | | Hibiscus Latent Singapore virus | | Descriptor: | Coat protein, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Tewary, S.K, Wong, S.M, Swaminathan, K. | | Deposit date: | 2010-10-22 | | Release date: | 2011-01-12 | | Last modified: | 2024-03-20 | | Method: | FIBER DIFFRACTION (3.5 Å) | | Cite: | Structure of Hibiscus latent Singapore virus by fiber diffraction: A non-conserved His122 contributes to coat protein stability
J.Mol.Biol., 2010
|
|
6E3E
 
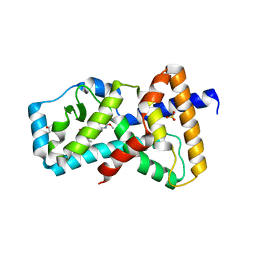 | | Structure of RORgt in complex with a novel inverse agonist. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-[(5R)-5-[(7-fluoro-1,1-dimethyl-2,3-dihydro-1H-inden-5-yl)carbamoyl]-2-methoxy-7,8-dihydro-1,6-naphthyridin-6(5H)-yl]-5-oxopentanoic acid, Nuclear receptor ROR-gamma, ... | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2018-07-13 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Retinoic Acid-Related Orphan Receptor gamma t (ROR gamma t) Agonist Structure-Based Functionality Switching Approach from In House ROR gamma t Inverse Agonist to ROR gamma t Agonist.
J.Med.Chem., 62, 2019
|
|
6E3G
 
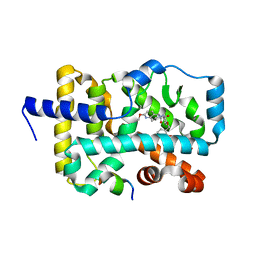 | | Structure of RORgt in complex with a novel agonist. | | Descriptor: | (5R)-6-acetyl-2-methoxy-N-{4-[(2-methoxyphenyl)methoxy]phenyl}-5,6,7,8-tetrahydro-1,6-naphthyridine-5-carboxamide, 1,2-ETHANEDIOL, Nuclear receptor ROR-gamma, ... | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2018-07-13 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Retinoic Acid-Related Orphan Receptor gamma t (ROR gamma t) Agonist Structure-Based Functionality Switching Approach from In House ROR gamma t Inverse Agonist to ROR gamma t Agonist.
J.Med.Chem., 62, 2019
|
|
6BR2
 
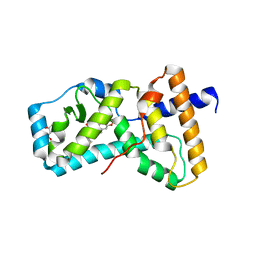 | | Structure of RORgt in complex with a novel isoquinoline inverse agonist. | | Descriptor: | (1R)-N-(4-tert-butyl-3-fluorophenyl)-6-methoxy-2-[(3-oxo-2,3-dihydro-1,2-oxazol-5-yl)acetyl]-1,2,3,4-tetrahydroisoquinoline-1-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Nuclear receptor ROR-gamma | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2017-11-29 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Discovery of [ cis-3-({(5 R)-5-[(7-Fluoro-1,1-dimethyl-2,3-dihydro-1 H-inden-5-yl)carbamoyl]-2-methoxy-7,8-dihydro-1,6-naphthyridin-6(5 H)-yl}carbonyl)cyclobutyl]acetic Acid (TAK-828F) as a Potent, Selective, and Orally Available Novel Retinoic Acid Receptor-Related Orphan Receptor gamma t Inverse Agonist.
J. Med. Chem., 61, 2018
|
|
6BR3
 
 | | Structure of RORgt in complex with a novel inverse agonist TAK-828. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Nuclear receptor ROR-gamma, {cis-3-[(5R)-5-[(7-fluoro-1,1-dimethyl-1H-inden-5-yl)carbamoyl]-2-methoxy-7,8-dihydro-1,6-naphthyridine-6(5H)-carbonyl]cyclobutyl}acetic acid | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2017-11-29 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of [ cis-3-({(5 R)-5-[(7-Fluoro-1,1-dimethyl-2,3-dihydro-1 H-inden-5-yl)carbamoyl]-2-methoxy-7,8-dihydro-1,6-naphthyridin-6(5 H)-yl}carbonyl)cyclobutyl]acetic Acid (TAK-828F) as a Potent, Selective, and Orally Available Novel Retinoic Acid Receptor-Related Orphan Receptor gamma t Inverse Agonist.
J. Med. Chem., 61, 2018
|
|
5H4S
 
 | |
3ASK
 
 | | Structure of UHRF1 in complex with histone tail | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3.3, ZINC ION | | Authors: | Arita, K, Sugita, K, Unoki, M, Hamamoto, R, Sekiyama, N, Tochio, H, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-25 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6J13
 
 | | Redox protein from Chlamydomonas reinhardtii | | Descriptor: | 2-cys peroxiredoxin | | Authors: | Charoenwattansatien, R, Zinzius, K, Tanaka, H, Hippler, M, Kurisu, G. | | Deposit date: | 2018-12-27 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Calcium sensing via EF-hand 4 enables thioredoxin activity in the sensor-responder protein calredoxin in the green algaChlamydomonas reinhardtii.
J.Biol.Chem., 295, 2020
|
|
3WKM
 
 | | The periplasmic PDZ tandem fragment of the RseP homologue from Aquifex aeolicus in complex with the Fab fragment | | Descriptor: | MOUSE IGG1-KAPPA FAB (HEAVY CHAIN), MOUSE IGG1-KAPPA FAB (LIGHT CHAIN), Putative zinc metalloprotease aq_1964 | | Authors: | Nogi, T, Tabata, S, Tamura-kawakami, K, Takagi, J. | | Deposit date: | 2013-10-28 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Structure-Based Model of Substrate Discrimination by a Noncanonical PDZ Tandem in the Intramembrane-Cleaving Protease RseP
Structure, 22, 2013
|
|
