7B1P
 
 | | Crystal Structure of Human BACE-1 in Complex with Compound 38a (NB-854) | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(3~{R},6~{R})-5-azanyl-3,6-dimethyl-6-(trifluoromethyl)-2~{H}-1,4-oxazin-3-yl]phenyl]-5-bromanyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.M, Wirth, E. | | Deposit date: | 2020-11-25 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Synthesis of the Potent, Selective, and Efficacious beta-Secretase (BACE1) Inhibitor NB-360.
J.Med.Chem., 64, 2021
|
|
7B1E
 
 | | BACE1 IN COMPLEX WITH compound 3 (NB-641) | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(4~{S})-2-azanyl-4-methyl-5,6-dihydro-1,3-thiazin-4-yl]phenyl]-5-bromanyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.M, Wirth, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Synthesis of the Potent, Selective, and Efficacious beta-Secretase (BACE1) Inhibitor NB-360.
J.Med.Chem., 64, 2021
|
|
7B1Q
 
 | | Crystal Structure of Human BACE-1 in Complex with Compound NB-360 (compound 54) | | Descriptor: | Beta-secretase 1, ~{N}-[3-[(3~{R},6~{R})-5-azanyl-3,6-dimethyl-6-(trifluoromethyl)-2~{H}-1,4-oxazin-3-yl]-4-fluoranyl-phenyl]-5-cyano-3-methyl-pyridine-2-carboxamide | | Authors: | Rondeau, J.M, Wirth, E. | | Deposit date: | 2020-11-25 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Synthesis of the Potent, Selective, and Efficacious beta-Secretase (BACE1) Inhibitor NB-360.
J.Med.Chem., 64, 2021
|
|
3QBH
 
 | | Structure based design, synthesis and SAR of cyclic hydroxyethylamine (HEA) BACE-1 inhibitors | | Descriptor: | (4S)-4-(2-hydroxy-5-{[(3S,4S,5R)-4-hydroxy-1,1-dioxido-5-{[3-(propan-2-yl)benzyl]amino}tetrahydro-2H-thiopyran-3-yl]methyl}benzyl)-3-propyl-1,3-oxazolidin-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M. | | Deposit date: | 2011-01-13 | | Release date: | 2011-03-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure based design, synthesis and SAR of cyclic hydroxyethylamine (HEA) BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5HXB
 
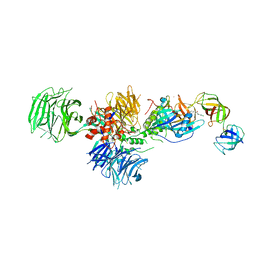 | | Cereblon in complex with DDB1, CC-885, and GSPT1 | | Descriptor: | 1-(3-chloro-4-methylphenyl)-3-({2-[(3S)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-5-yl}methyl)urea, DNA damage-binding protein 1, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, ... | | Authors: | Chamberlain, P.P, Matyskiela, M, Pagarigan, B. | | Deposit date: | 2016-01-30 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase.
Nature, 535, 2016
|
|
4CEL
 
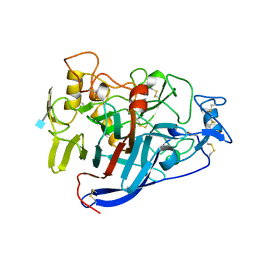 | | ACTIVE-SITE MUTANT D214N DETERMINED AT PH 6.0 WITH NO LIGAND BOUND IN THE ACTIVE SITE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1996-08-24 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 264, 1996
|
|
3CWE
 
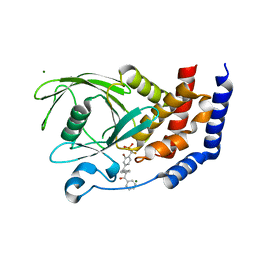 | | PTP1B in complex with a phosphonic acid inhibitor | | Descriptor: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1, [{2-bromo-4-[(2R)-3-oxo-2,3-diphenylpropyl]phenyl}(difluoro)methyl]phosphonic acid | | Authors: | Scapin, G, Han, Y, Kennedy, B.P. | | Deposit date: | 2008-04-21 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of [(3-bromo-7-cyano-2-naphthyl)(difluoro)methyl]phosphonic acid, a potent and orally active small molecule PTP1B inhibitor
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4BL2
 
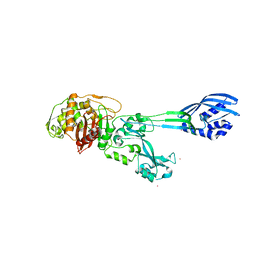 | |
6YN9
 
 | | MALT1(329-728) in complex with a sulfonamide containing compound | | Descriptor: | 5-[4-[(2,6-diethylphenyl)sulfamoyl]-3-methyl-phenyl]furan-3-carboxylic acid, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-04-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.558 Å) | | Cite: | Stabilizing Inactive Conformations of MALT1 as an Effective Approach to Inhibit Its Protease Activity
Advanced Therapeutics, 3, 2020
|
|
4US6
 
 | | New Crystal Form of Glucose Isomerase Grown in Short Peptide Supramolecular Hydrogels | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Gavira, J.A, Conejero-Muriel, M, Diaz-Mochon, J.J, Alvarez de Cienfuegos, L. | | Deposit date: | 2014-07-03 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Influence of the Chirality of Short Peptide Supramolecular Hydrogels in Protein Crystallogenesis.
Chem.Commun.(Camb.), 51, 2015
|
|
6MEP
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC3437 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine-protein kinase Mer, ... | | Authors: | Da, C, Zhang, D, Stashko, M.A, Cheng, A, Hunter, D, Norris-Drouin, J, Graves, L, Machius, M, Miley, M.J, DeRyckere, D, Earp, H.S, Graham, D.K, Frye, S.V, Wang, X, Kireev, D. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Data-Driven Construction of Antitumor Agents with Controlled Polypharmacology.
J.Am.Chem.Soc., 141, 2019
|
|
8C8T
 
 | | cryo-EM structure of BG505 SOSIP.664 HIV-1 Env trimer in complex with bNAbs EPTC112 and 3BNC117 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Baquero, E, Molinos-Albert, L, Mouquet, H. | | Deposit date: | 2023-01-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Anti-V1/V3-glycan broadly HIV-1 neutralizing antibodies in a post-treatment controller.
Cell Host Microbe, 31, 2023
|
|
6YN8
 
 | | Human MALT1(334-719) in complex with a tetrazole containing compound | | Descriptor: | 3-azanyl-3-methyl-~{N}-[(3~{R})-4-oxidanylidene-5-[[4-[2-(1~{H}-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]methyl]-2,3-dihydro-1,5-benzoxazepin-3-yl]butanamide, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M. | | Deposit date: | 2020-04-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Stabilizing Inactive Conformations of MALT1 as an Effective Approach to Inhibit Its Protease Activity
Advanced Therapeutics, 3, 2020
|
|
8CZ9
 
 | |
7PC2
 
 | | HIV-1 Env (BG505 SOSIP.664) in complex with the IgA bNAb 7-269 and the antibody 3BNC117. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC IgG Fab heavy chain, ... | | Authors: | Fernandez, I, Bontems, F, Pehau-Arnaudet, G, Rey, F. | | Deposit date: | 2021-08-03 | | Release date: | 2022-02-23 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Epitope convergence of broadly HIV-1 neutralizing IgA and IgG antibody lineages in a viremic controller.
J.Exp.Med., 219, 2022
|
|
5KHM
 
 | | The first BET bromodomain of BRD4 bound to compound 13 in a bivalent manner | | Descriptor: | (3~{R})-4-[2-[4-[1-(3-methoxy-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)piperidin-4-yl]phenoxy]ethyl]-1,3-dimethyl-piperazin-2-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Patel, J. | | Deposit date: | 2016-06-15 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Optimization of a Series of Bivalent Triazolopyridazine Based Bromodomain and Extraterminal Inhibitors: The Discovery of (3R)-4-[2-[4-[1-(3-Methoxy-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidyl]phenoxy]ethyl]-1,3-dimethyl-piperazin-2-one (AZD5153).
J.Med.Chem., 59, 2016
|
|
6PLI
 
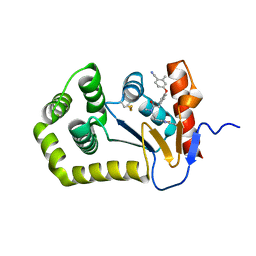 | | Crystal Structure of EcDsbA in a complex with purified oxadiazole 11 | | Descriptor: | 2-[4-(4-cyano-3-methylphenoxy)phenyl]-N-methyl-N-[2-(5-methyl-1,2,4-oxadiazol-3-yl)ethyl]acetamide, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-07-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PC9
 
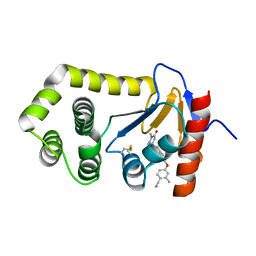 | | Crystal Structure of EcDsbA in a complex with purified methylpiperazinone 6 | | Descriptor: | 2-methyl-4-{4-[2-(4-methyl-3-oxopiperazin-1-yl)-2-oxoethyl]phenoxy}benzonitrile, TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-17 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiLX).
J.Med.Chem., 63, 2020
|
|
6PIQ
 
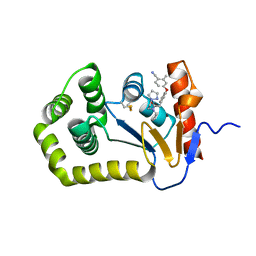 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product G6 (pyrazole 9) | | Descriptor: | 2-[4-(4-cyano-3-methylphenoxy)phenyl]-N-ethyl-N-[2-(1H-pyrazol-1-yl)ethyl]acetamide, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PG2
 
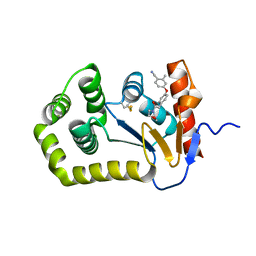 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product H5 (morpholine 8) | | Descriptor: | 2-methyl-4-{4-[2-(morpholin-4-yl)-2-oxoethyl]phenoxy}benzonitrile, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-23 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PBI
 
 | | Crystal Structure of EcDsbA in a complex with purified morpholine 8 | | Descriptor: | 2-methyl-4-{4-[2-(morpholin-4-yl)-2-oxoethyl]phenoxy}benzonitrile, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-13 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PGJ
 
 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product A5 (Morpholine carboxylic acid 7) | | Descriptor: | (3R)-4-{[4-(4-cyano-3-methylphenoxy)phenyl]acetyl}morpholine-3-carboxylic acid, (3S)-4-{[4-(4-cyano-3-methylphenoxy)phenyl]acetyl}morpholine-3-carboxylic acid, COPPER (II) ION, ... | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-24 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PG1
 
 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product F1 (methylpiperazinone 6) | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-4-{4-[2-(4-methyl-3-oxopiperazin-1-yl)-2-oxoethyl]phenoxy}benzonitrile, COPPER (II) ION, ... | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-23 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PDH
 
 | | Crystal Structure of EcDsbA in a complex with purified pyrazole 9 | | Descriptor: | 2-[4-(4-cyano-3-methylphenoxy)phenyl]-N-ethyl-N-[2-(1H-pyrazol-1-yl)ethyl]acetamide, TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-19 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PD7
 
 | | Crystal Structure of EcDsbA in a complex with purified morpholine carboxylic acid 7 | | Descriptor: | (3R)-4-{[4-(4-cyano-3-methylphenoxy)phenyl]acetyl}morpholine-3-carboxylic acid, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-18 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
