3N9M
 
 | | ceKDM7A from C.elegans, alone | | Descriptor: | FE (II) ION, Putative uncharacterized protein, ZINC ION | | Authors: | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | Deposit date: | 2010-05-31 | | Release date: | 2010-06-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
3N9P
 
 | | ceKDM7A from C.elegans, complex with H3K4me3K27me2 peptide and NOG | | Descriptor: | FE (II) ION, Histone H3 peptide, N-OXALYLGLYCINE, ... | | Authors: | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | Deposit date: | 2010-05-31 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
7O7T
 
 | | Structure of the PL6 family alginate lyase Patl3640 from Pseudoalteromonas atlantica T6c in complex with 4-deoxy-L-erythro-5-hexoseulose uronic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-deoxy-L-erythro-hex-5-ulosuronic acid, GLYCEROL, ... | | Authors: | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O84
 
 | | Structure of the PL6 family alginate lyase Pedsa0632 from Pseudopedobacter saltans in complex with substrate | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, Alginate lyase | | Authors: | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O7A
 
 | |
7O77
 
 | | Structure of the PL6 family alginate lyase Patl3640 from Pseudoalteromonas atlantica T6c | | Descriptor: | GLYCEROL, Poly(Beta-D-mannuronate) lyase | | Authors: | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.321 Å) | | Cite: | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O79
 
 | | Structure of the PL6 family polysaccharide lyase Pedsa3628 from Pseudopedobacter saltans | | Descriptor: | PHOSPHATE ION, Poly(Beta-D-mannuronate) lyase | | Authors: | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7O78
 
 | | Structure of the PL6 family chondroitinase B from Pseudopedobacter saltans, Pedsa3807 | | Descriptor: | Polysaccharide lyase from Pseudopedobacter saltans, Pedsa3807 | | Authors: | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
7X35
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 8A10 (CVB1-A:8A10) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
4Y60
 
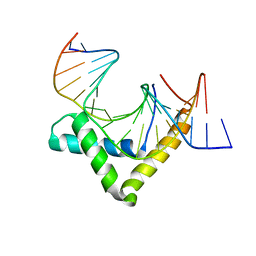 | | Structure of SOX18-HMG/PROX1-DNA | | Descriptor: | DNA (5'-D(*CP*AP*CP*TP*AP*GP*CP*AP*TP*TP*GP*TP*CP*TP*GP*GP*G)-3'), DNA (5'-D(*GP*CP*CP*CP*AP*GP*AP*CP*AP*AP*TP*GP*CP*TP*AP*GP*T)-3'), Transcription factor SOX-18 | | Authors: | Narasimhan, K, Prokoph, N, Kolatkar, P, Robinson, H, Jauch, R. | | Deposit date: | 2015-02-12 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and decoy-mediated inhibition of the SOX18/Prox1-DNA interaction.
Nucleic Acids Res., 44, 2016
|
|
7C2L
 
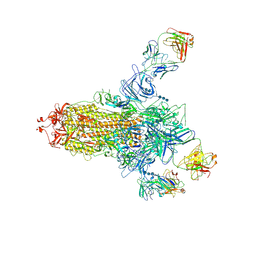 | | S protein of SARS-CoV-2 in complex bound with 4A8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Guo, Y.Y, Li, Y.N, Xia, L, Zhou, Q. | | Deposit date: | 2020-05-08 | | Release date: | 2020-07-01 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2.
Science, 369, 2020
|
|
7TDW
 
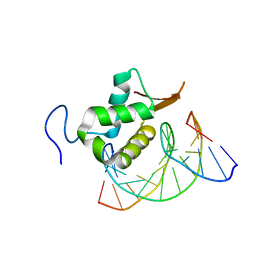 | | Structure of FOXP3-DNA complex | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*AP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*TP*AP*AP*AP*CP*AP*AP*AP*TP*TP*T)-3'), Forkhead box P3 | | Authors: | Leng, F, Hur, S. | | Deposit date: | 2022-01-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The transcription factor FoxP3 can fold into two dimerization states with divergent implications for regulatory T cell function and immune homeostasis.
Immunity, 55, 2022
|
|
7TDX
 
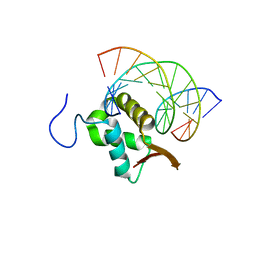 | | Structure of FOXP3-DNA complex | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*AP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*TP*AP*AP*AP*CP*AP*AP*AP*TP*TP*T)-3'), Forkhead box P3 | | Authors: | Leng, F, Hur, S. | | Deposit date: | 2022-01-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The transcription factor FoxP3 can fold into two dimerization states with divergent implications for regulatory T cell function and immune homeostasis.
Immunity, 55, 2022
|
|
4P6W
 
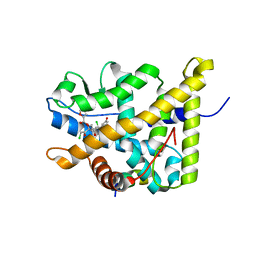 | | Crystal Structure of mometasone furoate-bound glucocorticoid receptor ligand binding domain | | Descriptor: | Glucocorticoid receptor, MOMETASONE FUROATE, Nuclear receptor coactivator 2 | | Authors: | He, Y, Zhou, X.E, Tolbert, W.D, Powell, K, Melcher, K, Xu, H.E. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structures and mechanism for the design of highly potent glucocorticoids.
Cell Res., 24, 2014
|
|
4P6X
 
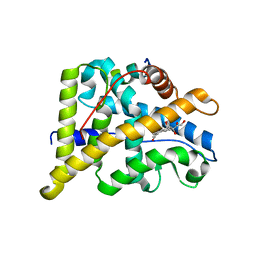 | | Crystal Structure of cortisol-bound glucocorticoid receptor ligand binding domain | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | He, Y, Zhou, X.E, Tolbert, W.D, Powell, K, Melcher, K, Xu, H.E. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and mechanism for the design of highly potent glucocorticoids.
Cell Res., 24, 2014
|
|
6IUR
 
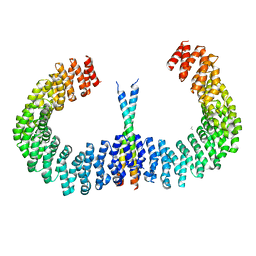 | | A phosphatase complex STRN3-PP2Aa | | Descriptor: | PP2A scaffolding subunit, PROPANE, Striatin-3 | | Authors: | Tang, Y, Zhou, Z.C. | | Deposit date: | 2018-11-30 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Selective Inhibition of STRN3-Containing PP2A Phosphatase Restores Hippo Tumor-Suppressor Activity in Gastric Cancer.
Cancer Cell, 38, 2020
|
|
8I3U
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 14B1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 14B1, Light chain of Fab 14B1, ... | | Authors: | Li, Z, Yu, F, Cao, S. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
8I3S
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 7B3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain od Fab 7B3, Light chain of Fab 7B3, ... | | Authors: | Li, Z, Yu, F, Cao, S, ZHao, H. | | Deposit date: | 2023-01-17 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
4O0U
 
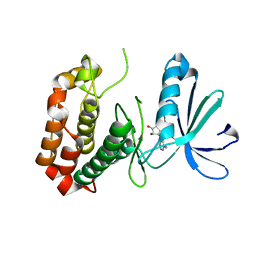 | |
3WY7
 
 | | Crystal structure of Mycobacterium smegmatis 7-Keto-8-aminopelargonic acid (KAPA) synthase BioF | | Descriptor: | 8-amino-7-oxononanoate synthase | | Authors: | Fan, S.H, Li, D.F, Wang, D.C, Chen, G.J, Zhang, X.E, Bi, L.J. | | Deposit date: | 2014-08-20 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of Mycobacterium smegmatis 7-keto-8-aminopelargonic acid (KAPA) synthase
Int.J.Biochem.Cell Biol., 58C, 2014
|
|
4O0S
 
 | |
4O0W
 
 | |
5UZ6
 
 | | RNA hairpin structure containing 2-MeImp-oligo analogue | | Descriptor: | RNA (25-MER), RNA (5'-D(*(8OS))-R(P*CP*AP*CP*CP*UP*CP*A)-3') | | Authors: | Zhang, W, Oh, S.S, Szostak, J.W. | | Deposit date: | 2017-02-24 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Rationale for the Enhanced Catalysis of Nonenzymatic RNA Primer Extension by a Downstream Oligonucleotide.
J. Am. Chem. Soc., 140, 2018
|
|
5V5O
 
 | |
5VCF
 
 | | RNA hairpin structure containing tetraloop/receptor motif, complexed with 2-MeImpG analogue | | Descriptor: | 5'-O-[(S)-hydroxy(4-methyl-1H-imidazol-5-yl)phosphoryl]guanosine, RNA (5'-R(*CP*UP*AP*UP*GP*CP*CP*UP*GP*CP*UP*G)-3'), RNA (5'-R(*CP*UP*GP*CP*UP*GP*GP*CP*UP*AP*AP*GP*GP*CP*CP*CP*GP*AP*AP*AP*GP*G)-3') | | Authors: | Zhang, W, Szostak, J.W. | | Deposit date: | 2017-03-31 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Rationale for the Enhanced Catalysis of Nonenzymatic RNA Primer Extension by a Downstream Oligonucleotide.
J. Am. Chem. Soc., 140, 2018
|
|
