2O88
 
 | |
8AH5
 
 | |
8AH6
 
 | |
8AH8
 
 | |
8AH4
 
 | |
8AH7
 
 | |
1NXD
 
 | | Crystal structure of MnMn Concanavalin A | | Descriptor: | AZIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lopez-Jaramillo, F.J, Gonzalez-Ramirez, L.A, Albert, A, Santoyo-Gonzalez, F, Vargas-Berenguel, A, Otalora, F. | | Deposit date: | 2003-02-10 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of concanavalin A at pH 8: bound solvent and crystal contacts.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6F84
 
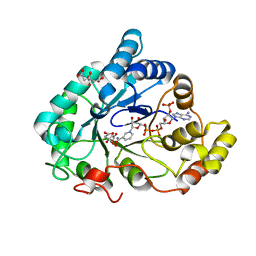 | | AKR1B1 at 2.55 MGy radiation dose. | | Descriptor: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Castellvi, A, Juanhuix, J. | | Deposit date: | 2017-12-12 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation.
Sci Rep, 9, 2019
|
|
6F8O
 
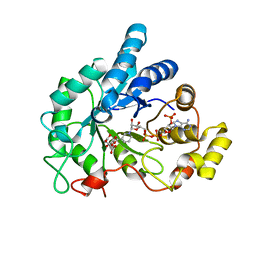 | | AKR1B1 at 3.45 MGy radiation dose. | | Descriptor: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Castellvi, A, Juanhuix, J. | | Deposit date: | 2017-12-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation.
Sci Rep, 9, 2019
|
|
6F81
 
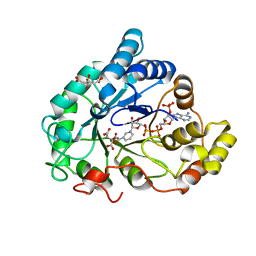 | | AKR1B1 at 0.75 MGy radiation dose. | | Descriptor: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Castellvi, A, Juanhuix, J. | | Deposit date: | 2017-12-12 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation.
Sci Rep, 9, 2019
|
|
6F7R
 
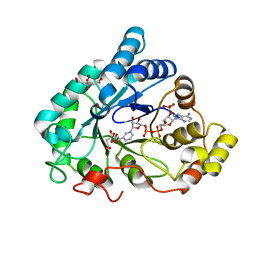 | | AKR1B1 at 0.03 MGy radiation dose. | | Descriptor: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Castellvi, A, Juanhuix, J. | | Deposit date: | 2017-12-11 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation.
Sci Rep, 9, 2019
|
|
6F82
 
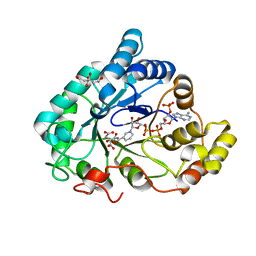 | | AKR1B1 at 1.65 MGy radiation dose. | | Descriptor: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Castellvi, A, Juanhuix, J. | | Deposit date: | 2017-12-12 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation.
Sci Rep, 9, 2019
|
|
3HG6
 
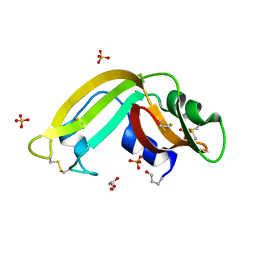 | | Crystal Structure of the Recombinant Onconase from Rana pipiens | | Descriptor: | GLYCEROL, Onconase, SULFATE ION | | Authors: | Camara-Artigas, A, Gavira, J.A, Casares-Atienza, S, Weininger, U, Balbach, J, Garcia-Mira, M.M. | | Deposit date: | 2009-05-13 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three-state thermal unfolding of onconase.
Biophys.Chem., 159, 2011
|
|
3I9Q
 
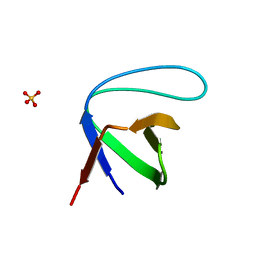 | |
8AND
 
 | | Domain swapped dimer of smolstatin (stefin) from Sphaerospora molnari | | Descriptor: | 1,2-ETHANEDIOL, Smolstatin | | Authors: | Havlickova, P, Bartosova-Sojkova, P, Sojka, D, Kascakova, B, Gavira, J.A, Kuta Smatanova, I. | | Deposit date: | 2022-08-05 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Domain swapped dimer of smolstatin (stefin) from Sphaerospora molnari
To Be Published
|
|
3EG0
 
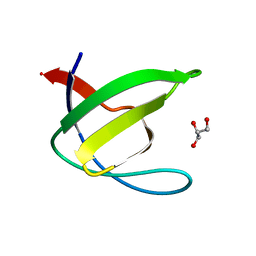 | | Crystal structure of the N114T mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-10 | | Release date: | 2009-09-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
3NGP
 
 | |
3EG2
 
 | | Crystal structure of the N114Q mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-10 | | Release date: | 2009-09-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
3EG1
 
 | |
3EGU
 
 | | Crystal structure of the N114A mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1, SULFATE ION | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-11 | | Release date: | 2009-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
3EG3
 
 | | Crystal structure of the N114A mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-10 | | Release date: | 2009-09-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
4J9E
 
 | |
4J9D
 
 | |
4JJB
 
 | |
4J9B
 
 | |
