8AV1
 
 | | Crystal structure of GSK3 beta (GSK3b) in complex with CD7. | | Descriptor: | 1,2-ETHANEDIOL, 2-pyridin-3-yl-8-thiomorpholin-4-yl-[1,3]oxazolo[5,4-f]quinoxaline, Glycogen synthase kinase-3 beta, ... | | Authors: | Chaikuad, A, Mongin, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Oxazolo[5,4-f]quinoxaline-type selective inhibitors of glycogen synthase kinase-3 alpha (GSK-3 alpha ): Development and impact on temozolomide treatment of glioblastoma cells.
Bioorg.Chem., 134, 2023
|
|
8AUZ
 
 | | Crystal structure of GSK3 beta (GSK3b) in complex with FL291. | | Descriptor: | 8-morpholin-4-yl-2-pyridin-3-yl-[1,3]oxazolo[5,4-f]quinoxaline, Glycogen synthase kinase-3 beta, SULFATE ION | | Authors: | Chaikuad, A, Mongin, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Oxazolo[5,4-f]quinoxaline-type selective inhibitors of glycogen synthase kinase-3 alpha (GSK-3 alpha ): Development and impact on temozolomide treatment of glioblastoma cells.
Bioorg.Chem., 134, 2023
|
|
7NCF
 
 | | Crystal structure of HIPK2 in complex with MU135 (compound 21e) | | Descriptor: | 3-(4-Tert-butylphenyl)-5-(1H-pyrazol-4-yl)furo[3,2-b]pyridine, Homeodomain-interacting protein kinase 2 | | Authors: | Chaikuad, A, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Highly selective inhibitors of protein kinases CLK and HIPK with the furo[3,2-b]pyridine core.
Eur.J.Med.Chem., 215, 2021
|
|
8PP0
 
 | | Crystal structure of Retinoic Acid Receptor alpha (RXRA) in complexed with JP147 | | Descriptor: | 3-[4-[2,3-dihydro-1H-inden-4-yl(methyl)amino]-6-(trifluoromethyl)pyrimidin-2-yl]oxypropanoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chaikuad, A, Pollinger, J, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-07-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of a Highly Potent Partial RXR Agonist with Superior Physicochemical Properties.
J.Med.Chem., 67, 2024
|
|
7Z71
 
 | | Crystal structure of p63 DBD in complex with darpin C14 | | Descriptor: | Darpin C14, Isoform 4 of Tumor protein 63, ZINC ION | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
7Z73
 
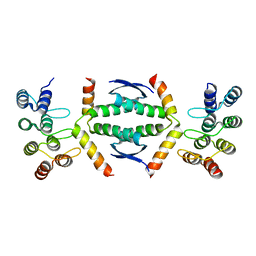 | | Crystal structure of p63 tetramerization domain in complex with darpin 8F1 | | Descriptor: | Darpin 8F1, Isoform 2 of Tumor protein 63 | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
7Z72
 
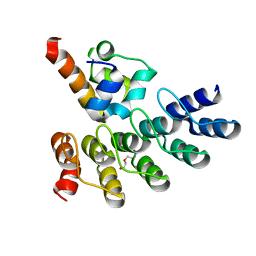 | | Crystal structure of p63 SAM in complex with darpin A5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Darpin A5, Isoform 9 of Tumor protein 63 | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
8ATZ
 
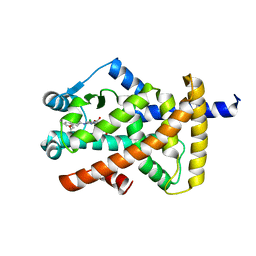 | | Crystal structure of PPAR gamma (PPARG) in complex with SA112 (compound 2). | | Descriptor: | 2-[4-chloranyl-6-[[3-(2-phenylethoxy)phenyl]amino]pyrimidin-2-yl]sulfanylethanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Arifi, S, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-08-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
8ATY
 
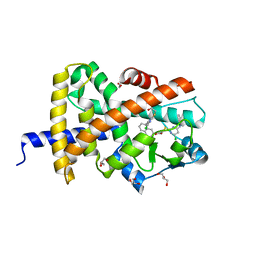 | | Crystal structure of PPAR gamma (PPARG) in complex with JP85 (compound 1). | | Descriptor: | 2-[4-chloranyl-6-(5,6,7,8-tetrahydronaphthalen-1-ylamino)pyrimidin-2-yl]sulfanylethanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Pollinger, J, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-08-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
8CPH
 
 | | Crystal structure of PPAR gamma (PPARG) in complex with WY-14643 (inactive form) | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
8CPI
 
 | | Crystal structure of PPAR gamma (PPARG) in complex with WY-14643 | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
8CPJ
 
 | | Crystal structure of PPAR gamma (PPARG) in an inactive form | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
5HTB
 
 | | Crystal structure of haspin (GSG2) in complex with bisubstrate inhibitor ARC-3353. | | Descriptor: | (2R,3S,4R,5R,6R)-6-((1R,2R,3S,4R,6S)-4,6-DIAMINO-2,3-DIHYDROXYCYCLOHEXYLOXY)-5-AMINO-2-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-3,4-DIOL, (3R)-4-amino-3-{[6-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]carbonyl}amino)hexanoyl]amino}-4-oxobutanoic acid (non-preferred name), (4R)-2-METHYLPENTANE-2,4-DIOL, ... | | Authors: | Chaikuad, A, Heroven, C, Lavogina, D, Kestav, K, Uri, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-01-26 | | Release date: | 2016-05-11 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-crystal structures of the protein kinase haspin with bisubstrate inhibitors.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7ZHP
 
 | | Crystal structure of TTBK1 in complex with compound 9 (7-005) | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-azanyl-3,5,12-triazatetracyclo[9.7.0.0^{2,7}.0^{13,18}]octadeca-1(11),2,4,6,13(18),14,16-heptaen-16-yl)-3-ethyl-pent-1-yn-3-ol, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Axtman, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of tau tubulin kinases (TTBK1 and TTBK2) impacts ciliogenesis.
Sci Rep, 13, 2023
|
|
5HTC
 
 | | Crystal structure of haspin (GSG2) in complex with bisubstrate inhibitor ARC-3372 | | Descriptor: | (2R)-2-{[6-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]carbonyl}amino)hexanoyl]amino}butanedioic acid (non-preferred name), (4S)-2-METHYL-2,4-PENTANEDIOL, ARC-3372 INHIBITOR, ... | | Authors: | Chaikuad, A, Heroven, C, Lavogina, D, Kestav, K, Uri, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-01-26 | | Release date: | 2016-03-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Co-crystal structures of the protein kinase haspin with bisubstrate inhibitors.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7ZHN
 
 | | Crystal structure of TTBK1 in complex with AMG28 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-amino-5,6,7,8-tetrahydropyrimido[4',5':3,4]cyclohepta[1,2-b]indol-11-yl)-2-methylbut-3-yn-2-ol, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Axtman, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Modulation of tau tubulin kinases (TTBK1 and TTBK2) impacts ciliogenesis.
Sci Rep, 13, 2023
|
|
7ZHQ
 
 | | Crystal structure of TTBK1 in complex with compound 10 (7-009) | | Descriptor: | (3~{S})-1-(4-azanyl-3,5,12-triazatetracyclo[9.7.0.0^{2,7}.0^{13,18}]octadeca-1(11),2,4,6,13(18),14,16-heptaen-16-yl)-3-methyl-pent-1-yn-3-ol, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Axtman, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of tau tubulin kinases (TTBK1 and TTBK2) impacts ciliogenesis.
Sci Rep, 13, 2023
|
|
7ZHO
 
 | | Crystal structure of TTBK1 in complex with compound 3 (7-001) | | Descriptor: | 1,2-ETHANEDIOL, 4-[3-(2-azanylpyrimidin-4-yl)-1~{H}-indol-5-yl]-2-methyl-but-3-yn-2-ol, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Axtman, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Modulation of tau tubulin kinases (TTBK1 and TTBK2) impacts ciliogenesis.
Sci Rep, 13, 2023
|
|
9GLQ
 
 | | Crystal structure of p73 tetramerisation domain in complex with darpins 1800 | | Descriptor: | COBALT (II) ION, Darpins 1800, GLYCEROL, ... | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-08-27 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DARPins as a novel tool to detect and degrade p73.
Cell Death Dis, 15, 2024
|
|
9FLC
 
 | | Crystal structure of haspin (GSG2) in complex with MU1668 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(1-methylpyrazol-3-yl)-3-pyridin-4-yl-thieno[3,2-b]pyridine, GLYCEROL, ... | | Authors: | Chaikuad, A, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-06-04 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Thieno[3,2-b]pyridine: Attractive scaffold for highly selective inhibitors of underexplored protein kinases with variable binding mode.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9FLB
 
 | | Crystal structure of haspin (GSG2) in complex with MU1464 | | Descriptor: | 5-(1-methylpyrazol-4-yl)-3-pyridin-4-yl-thieno[3,2-b]pyridine, SODIUM ION, Serine/threonine-protein kinase haspin | | Authors: | Chaikuad, A, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-06-04 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Thieno[3,2-b]pyridine: Attractive scaffold for highly selective inhibitors of underexplored protein kinases with variable binding mode.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8C7Y
 
 | | Crystal structure of BRAF V600E in complex with a hybrid compound 6 | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, Serine/threonine-protein kinase B-raf, ... | | Authors: | Chaikuad, A, Bonnet, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-01-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design, synthesis and characterisation of a novel type II B-RAF paradox breaker inhibitor.
Eur.J.Med.Chem., 250, 2023
|
|
8C7X
 
 | | Crystal structure of BRAF in complex with a hybrid compound 6 | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Serine/threonine-protein kinase B-raf, ... | | Authors: | Chaikuad, A, Bonnet, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-01-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design, synthesis and characterisation of a novel type II B-RAF paradox breaker inhibitor.
Eur.J.Med.Chem., 250, 2023
|
|
5J1W
 
 | | Crystal structure of human CLK1 in complex with pyrido[3,4-g]quinazoline derivative ZW31 (compound 14) | | Descriptor: | Dual specificity protein kinase CLK1, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Esvan, Y.J, Zeinyeh, W, Boibessot, T, Nauton, L, Thery, V, Loaec, N, Meijer, L, Giraud, F, Moreau, P, Anizon, F, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery of pyrido[3,4-g]quinazoline derivatives as CMGC family protein kinase inhibitors: Design, synthesis, inhibitory potency and X-ray co-crystal structure.
Eur.J.Med.Chem., 118, 2016
|
|
4YU2
 
 | | Crystal structure of DYRK1A with harmine-derivatized AnnH-75 inhibitor | | Descriptor: | (1-chloro-7-methoxy-9H-beta-carbolin-9-yl)acetonitrile, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ... | | Authors: | Chaikuad, A, Wurzlbauer, A, Nowak, R, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bracher, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | How to Separate Kinase Inhibition from Undesired Monoamine Oxidase A Inhibition-The Development of the DYRK1A Inhibitor AnnH75 from the Alkaloid Harmine.
Molecules, 25, 2020
|
|
