5N2B
 
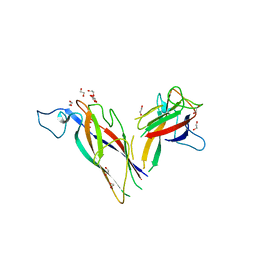 | |
1KR7
 
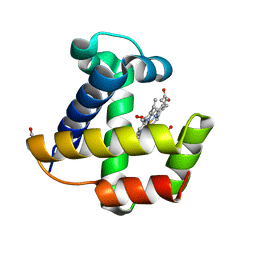 | | Crystal structure of the nerve tissue mini-hemoglobin from the nemertean worm Cerebratulus lacteus | | Descriptor: | ACETATE ION, Neural globin, OXYGEN MOLECULE, ... | | Authors: | Pesce, A, Nardini, M, Dewilde, S, Geuens, E, Yamauchi, k, Ascenzi, P, Riggs, A.F, Moens, L, Bolognesi, M. | | Deposit date: | 2002-01-09 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 109 residue nerve tissue minihemoglobin from Cerebratulus lacteus highlights striking structural plasticity of the alpha-helical globin fold
Structure, 10, 2002
|
|
1OAL
 
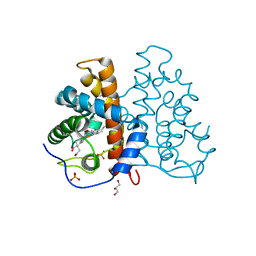
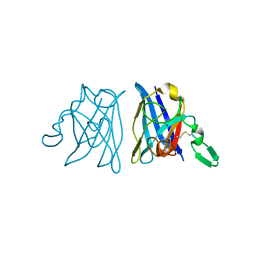 | | Active site copper and zinc ions modulate the quaternary structure of prokaryotic Cu,Zn superoxide dismutase | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Cioni, P, Pesce, A, Rocca, B.M.D, Castellifalconiparrilli, L, Bolognesi, M, Strambini, G, Desideri, A. | | Deposit date: | 2003-01-15 | | Release date: | 2003-02-27 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active-Site Copper and Zinc Ions Modulate the Quaternary Structure of Prokaryotic Cu,Zn Superoxide Dismutase
J.Mol.Biol., 326, 2003
|
|
2VEB
 
 | | High resolution structure of protoglobin from Methanosarcina acetivorans C2A | | Descriptor: | GLYCEROL, OXYGEN MOLECULE, PHOSPHATE ION, ... | | Authors: | Nardini, M, Pesce, A, Thijs, L, Saito, J.A, Dewilde, S, Alam, M, Ascenzi, P, Coletta, M, Ciaccio, C, Moens, L, Bolognesi, M. | | Deposit date: | 2007-10-18 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Archaeal Protoglobin Structure Indicates New Ligand Diffusion Paths and Modulation of Haem-Reactivity.
Embo Rep., 9, 2008
|
|
2VEE
 
 | | Structure of protoglobin from Methanosarcina acetivorans C2A | | Descriptor: | PROTOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nardini, M, Pesce, A, Thijs, L, Saito, J.A, Dewilde, S, Alam, M, Ascenzi, P, Coletta, M, Ciaccio, C, Moens, L, Bolognesi, M. | | Deposit date: | 2007-10-22 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Archaeal Protoglobin Structure Indicates New Ligand Diffusion Paths and Modulation of Haem-Reactivity.
Embo Rep., 9, 2008
|
|
4MTZ
 
 | | Structure of XIAP-BIR1 in complex with NF023 | | Descriptor: | 8-({3-[({3-[(4,6,8-trisulfonaphthalen-1-yl)carbamoyl]phenyl}carbamoyl)amino]benzoyl}amino)naphthalene-1,3,5-trisulfonic acid, E3 ubiquitin-protein ligase XIAP, SULFATE ION, ... | | Authors: | Cossu, F, Milani, M, Grassi, S, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein-protein interactions in nfkb pathway: novel tools to promote apoptosis in cancer therapy
To be Published
|
|
5MUD
 
 | | Crystal structure of an amyloidogenic light chain dimer H6 | | Descriptor: | light chain dimer,IGL@ protein | | Authors: | Oberti, L, Bacarizo, J, Maritan, M, Rognoni, P, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
1FLP
 
 | | STRUCTURE OF THE SULFIDE-REACTIVE HEMOGLOBIN FROM THE CLAM LUCINA PECTINATA: CRYSTALLOGRAPHIC ANALYSIS AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | HEMOGLOBIN I (AQUO MET), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Rizzi, M, Wittenberg, J.B, Ascenzi, P, Fasano, M, Coda, A, Bolognesi, M. | | Deposit date: | 1994-05-16 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the sulfide-reactive hemoglobin from the clam Lucina pectinata. Crystallographic analysis at 1.5 A resolution.
J.Mol.Biol., 244, 1994
|
|
2V2K
 
 | | THE CRYSTAL STRUCTURE OF FDXA, A 7FE FERREDOXIN FROM MYCOBACTERIUM SMEGMATIS | | Descriptor: | ACETATE ION, FE3-S4 CLUSTER, FERREDOXIN | | Authors: | Ricagno, S, de Rosa, M, Aliverti, A, Zanetti, G, Bolognesi, M. | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Fdxa, a 7Fe Ferredoxin from Mycobacterium Smegmatis.
Biochem.Biophys.Res.Commun., 360, 2007
|
|
5MUH
 
 | | Crystal structure of an amyloidogenic light chain dimer H7 | | Descriptor: | light chain dimer | | Authors: | Oberti, L, Rognoni, P, Russo, R, Maritan, M, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
2WTG
 
 | | High resolution 3D structure of C.elegans globin-like protein GLB-1 | | Descriptor: | GLOBIN-LIKE PROTEIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Geuens, E, Hoogewijs, D, Nardini, M, Vinck, E, Pesce, A, Kiger, L, Fago, A, Tilleman, L, De Henau, S, Marden, M, Weber, R.E, Van Doorslaer, S, Vanfleteren, J, Moens, L, Bolognesi, M, Dewilde, S. | | Deposit date: | 2009-09-16 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Globin-Like Proteins in Caenorhabditis Elegans: In Vivo Localization, Ligand Binding and Structural Properties.
Bmc Biochem., 11, 2010
|
|
2WTH
 
 | | Low resolution 3D structure of C.elegans globin-like protein (GLB-1): P3121 crystal form | | Descriptor: | GLOBIN-LIKE PROTEIN, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Geuens, E, Hoogewijs, D, Nardini, M, Vinck, E, Pesce, A, Kiger, L, Fago, A, Tilleman, L, De Henau, S, Marden, M, Weber, R.E, Van Doorslaer, S, Vanfleteren, J, Moens, L, Bolognesi, M, Dewilde, S. | | Deposit date: | 2009-09-16 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Globin-Like Proteins in Caenorhabditis Elegans: In Vivo Localization, Ligand Binding and Structural Properties.
Bmc Biochem., 11, 2010
|
|
5M6A
 
 | | Crystal structure of cardiotoxic Bence-Jones light chain dimer H9 | | Descriptor: | Bence-Jones light chain, GLYCEROL, PHOSPHATE ION | | Authors: | Oberti, L, Rognoni, P, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-24 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
1SDY
 
 | | STRUCTURE SOLUTION AND MOLECULAR DYNAMICS REFINEMENT OF THE YEAST CU,ZN ENZYME SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Djinovic, K, Gatti, G, Coda, A, Antolini, L, Pelosi, G, Desideri, A, Falconi, M, Marmocchi, F, Rotilio, G, Bolognesi, M. | | Deposit date: | 1991-06-14 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure solution and molecular dynamics refinement of the yeast Cu,Zn enzyme superoxide dismutase.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
5MTL
 
 | | Crystal structure of an amyloidogenic light chain | | Descriptor: | light chain dimer,IGL@ protein,IGL@ protein | | Authors: | Oberti, L, Rognoni, P, Russo, R, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-10 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5MVG
 
 | | Crystal structure of non-amyloidogenic light chain dimer M7 | | Descriptor: | GLYCEROL, light chain dimer | | Authors: | Oberti, L, Rognoni, P, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-16 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
4RMS
 
 | |
4RMT
 
 | | Crystal structure of the D98N Beta-2 Microglobulin mutant | | Descriptor: | ACETATE ION, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | de Rosa, M, Bolognesi, M, Ricagno, S. | | Deposit date: | 2014-10-22 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.242 Å) | | Cite: | Decoding the Structural Bases of D76N 2-Microglobulin High Amyloidogenicity through Crystallography and Asn-Scan Mutagenesis.
Plos One, 10, 2015
|
|
4RMR
 
 | |
4RMQ
 
 | |
5M76
 
 | | Crystal structure of cardiotoxic Bence-Jones light chain dimer H10 | | Descriptor: | BROMIDE ION, light chain dimer | | Authors: | Oberti, L, Rognoni, P, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-26 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5M6I
 
 | | Crystal structure of non-cardiotoxic Bence-Jones light chain dimer M8 | | Descriptor: | SODIUM ION, light chain dimer | | Authors: | Oberti, L, Rognoni, P, Russo, R, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-25 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
7ZTH
 
 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the open conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-05-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZLA
 
 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the half-closed conformation | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Savino, C, Exertier, C, Bolognesi, M, Chaves Sanjuan, A. | | Deposit date: | 2022-04-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7ZN5
 
 | | Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C2 symmetry. | | Descriptor: | DNA (48-MER), PLP-dependent aminotransferase family protein | | Authors: | Freda, I, Montemiglio, L.C, Tramonti, A, Contestabile, R, Vallone, B, Exertier, C, Savino, C, Chaves Sanjuan, A, Bolognesi, M. | | Deposit date: | 2022-04-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
