2MIV
 
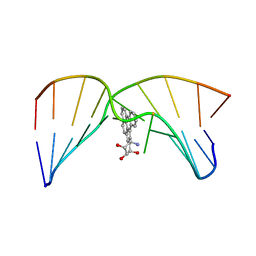 | | NMR studies of N2-guanine adducts derived from the tumorigen dibenzo[a,l]pyrene in DNA: Impact of adduct stereochemistry, size, and local DNA structure on solution conformations | | Descriptor: | (11R,12R,13R)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA_(5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA_(5'-D(*GP*GP*TP*AP*GP*GP*AP*TP*GP*G)-3') | | Authors: | Rodriguez, F.A, Liu, Z, Lin, C.H, Ding, S, Cai, Y, Kolbanovskiy, A, Kolbanovskiy, M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Studies of an N(2)-Guanine Adduct Derived from the Tumorigen Dibenzo[a,l]pyrene in DNA: Impact of Adduct Stereochemistry, Size, and Local DNA Sequence on Solution Conformations.
Biochemistry, 53, 2014
|
|
7P8E
 
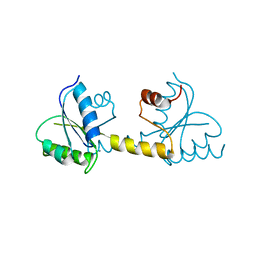 | | Crystal structure of the Receiver domain of M. truncatula cytokinin receptor MtCRE1 | | Descriptor: | CALCIUM ION, Receiver domain of histidine kinase | | Authors: | Tran, L.H, Urbanowicz, A, Jasinski, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3D Domain Swapping Dimerization of the Receiver Domain of Cytokinin Receptor CRE1 From Arabidopsis thaliana and Medicago truncatula .
Front Plant Sci, 12, 2021
|
|
2KE1
 
 | | Molecular Basis of non-modified histone H3 tail Recognition by the First PHD Finger of Autoimmune Regulator | | Descriptor: | Autoimmune regulator, H3K4me0, ZINC ION | | Authors: | Chignola, F, Gaetani, M, Rebane, A, Org, T, Mollica, L, Zucchelli, C, Spitaleri, A, Mannella, V, Peterson, P, Musco, G. | | Deposit date: | 2009-01-22 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the first PHD finger of autoimmune regulator in complex with non-modified histone H3 tail reveals the antagonistic role of H3R2 methylation
Nucleic Acids Res., 37, 2009
|
|
1AXL
 
 | | SOLUTION CONFORMATION OF THE (-)-TRANS-ANTI-[BP]DG ADDUCT OPPOSITE A DELETION SITE IN DNA DUPLEX D(CCATC-[BP]G-CTACC)D(GGTAG--GATGG), NMR, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CCATC-[BP]G-CTACC)D(GGTAG--GATGG) | | Authors: | Feng, B, Gorin, A.A, Kolbanovskiy, A, Hingerty, B.E, Geacintov, N.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-16 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of the (-)-trans-anti-[BP]dG adduct opposite a deletion site in a DNA duplex: intercalation of the covalently attached benzo[a]pyrene into the helix with base displacement of the modified deoxyguanosine into the minor groove.
Biochemistry, 36, 1997
|
|
6BMN
 
 | | Structure of human DHHC20 palmitoyltransferase, space group P63 | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, ZINC ION, ... | | Authors: | Rana, M.S, Lee, C.-J, Banerjee, A. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-24 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Fatty acyl recognition and transfer by an integral membraneS-acyltransferase.
Science, 359, 2018
|
|
6CMV
 
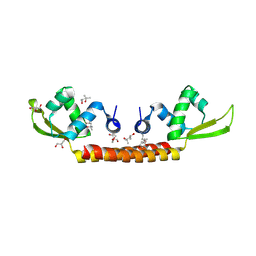 | | Crystal structure of Archaeal Biofilm Regulator (AbfR2) from Sulfolobus acidocaldarius | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Transcriptional regulator Lrs14-like protein | | Authors: | Essen, L.-O, Vogt, M.S, Banerjee, A. | | Deposit date: | 2018-03-06 | | Release date: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of an Lrs14-like archaeal biofilm regulator from Sulfolobus acidocaldarius.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6BML
 
 | | Structure of human DHHC20 palmitoyltransferase, irreversibly inhibited by 2-bromopalmitate | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, PALMITIC ACID, PHOSPHATE ION, ... | | Authors: | Rana, M.S, Lee, C.-J, Banerjee, A. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-24 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Fatty acyl recognition and transfer by an integral membraneS-acyltransferase.
Science, 359, 2018
|
|
6BMM
 
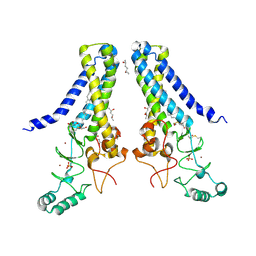 | | Structure of human DHHC20 palmitoyltransferase, space group P21 | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,5S)-hexane-2,5-diol, PHOSPHATE ION, ... | | Authors: | Rana, M.S, Lee, C.-J, Banerjee, A. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-24 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fatty acyl recognition and transfer by an integral membraneS-acyltransferase.
Science, 359, 2018
|
|
5TO6
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Bandyopadhyay, A, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
1Y9H
 
 | | Methylation of cytosine at C5 in a CpG sequence context causes a conformational switch of a benzo[a]pyrene diol epoxide-N2-guanine adduct in DNA from a minor groove alignment to intercalation with base displacement | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 5'-D(*CP*CP*AP*TP*(5CM)P*(BPG)P*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Zhang, N, Lin, C, Huang, X, Kolbanovskiy, A, Hingerty, B.E, Amin, S, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 2004-12-15 | | Release date: | 2005-03-22 | | Last modified: | 2024-04-24 | | Method: | SOLUTION NMR | | Cite: | Methylation of cytosine at C5 in a CpG sequence context causes a conformational switch of a benzo[a]pyrene diol epoxide-N2-guanine adduct in DNA from a minor groove alignment to intercalation with base displacement.
J.Mol.Biol., 346, 2005
|
|
7KHM
 
 | | Crystal structure of hDHHS20 bound to palmitoyl CoA | | Descriptor: | Isoform 4 of Palmitoyltransferase ZDHHC20, PHOSPHATE ION, Palmitoyl-CoA, ... | | Authors: | Lee, C.-J, Banerjee, A. | | Deposit date: | 2020-10-21 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Bivalent recognition of fatty acyl-CoA by a human integral membrane palmitoyltransferase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7KW9
 
 | | NMR Structure of a tRNA 2'-phosphotransferase from Runella slithyformis in complex with NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, tRNA 2'-phosphotransferase | | Authors: | Alphonse, S, Dantuluri, S, Banerjee, A, Shuman, S, Ghose, R. | | Deposit date: | 2020-11-30 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of Runella slithyformis RNA 2'-phosphotransferase Tpt1 provide insights into NAD+ binding and specificity.
Nucleic Acids Res., 49, 2021
|
|
7KW8
 
 | | NMR Structure of a tRNA 2'-phosphotransferase from Runella slithyformis | | Descriptor: | tRNA 2'-phosphotransferase | | Authors: | Alphonse, S, Dantuluri, S, Banerjee, A, Shuman, S, Ghose, R. | | Deposit date: | 2020-11-30 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of Runella slithyformis RNA 2'-phosphotransferase Tpt1 provide insights into NAD+ binding and specificity.
Nucleic Acids Res., 49, 2021
|
|
3QKM
 
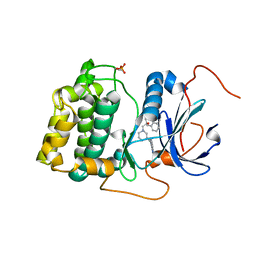 | | Spirocyclic sulfonamides as AKT inhibitors | | Descriptor: | N-(2-ethoxyethyl)-N-{(2S)-2-hydroxy-3-[(5R)-2-(quinazolin-4-yl)-2,7-diazaspiro[4.5]dec-7-yl]propyl}-2,6-dimethylbenzenesulfonamide, RAC-alpha serine/threonine-protein kinase | | Authors: | Xu, R, Banka, A, Blake, J.F, Mitchell, I.S, Wallace, E.M, Gloor, S.L, Martinson, M, Risom, T, Gross, S.D, Morales, T, Vigers, G.P.A, Brandhuber, B.J, Skelton, N.J. | | Deposit date: | 2011-02-01 | | Release date: | 2011-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of spirocyclic sulfonamides as potent Akt inhibitors with exquisite selectivity against PKA.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2LNC
 
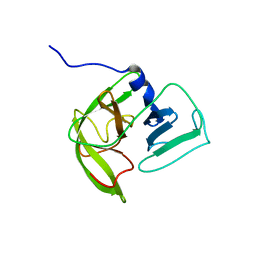 | | Solution NMR structure of Norwalk virus protease | | Descriptor: | 3C-like protease | | Authors: | Takahashi, D, Hiromasa, Y, Kim, Y, Anbanandam, A, Chang, K, Prakash, O. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamics characterization of norovirus protease.
Protein Sci., 22, 2013
|
|
1RRQ
 
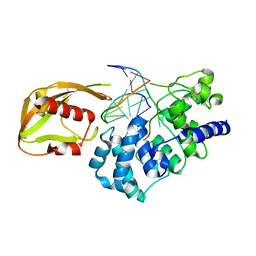 | | MutY adenine glycosylase in complex with DNA containing an A:oxoG pair | | Descriptor: | 5'-D(*TP*GP*TP*CP*CP*AP*AP*GP*TP*CP*T)-3', 5'-D(AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3', CALCIUM ION, ... | | Authors: | Fromme, J.C, Banerjee, A, Huang, S.J, Verdine, G.L. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase
Nature, 427, 2004
|
|
1RRS
 
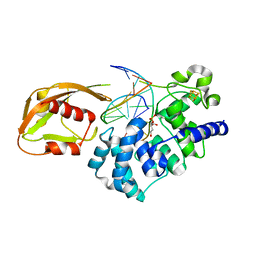 | | MutY adenine glycosylase in complex with DNA containing an abasic site | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*T)-3', CALCIUM ION, ... | | Authors: | Fromme, J.C, Banerjee, A, Huang, S.J, Verdine, G.L. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase
Nature, 427, 2004
|
|
1VRL
 
 | | MutY adenine glycosylase in complex with DNA and soaked adenine free base | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*T)-3', ADENINE, ... | | Authors: | Fromme, J.C, Banerjee, A, Huang, S.J, Verdine, G.L. | | Deposit date: | 2005-03-08 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase
Nature, 427, 2004
|
|
1AU8
 
 | | HUMAN CATHEPSIN G | | Descriptor: | CATHEPSIN G, N-(3-carboxypropanoyl)-L-valyl-N-[(1R)-5-amino-1-phosphonopentyl]-L-prolinamide | | Authors: | Medrano, F.J, Bode, W, Banbula, A, Potempa, J. | | Deposit date: | 1997-09-12 | | Release date: | 1998-10-14 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HUMAN CATHEPSIN G
to be published
|
|
2NOF
 
 | | Structure of Q315F human 8-oxoguanine glycosylase proximal crosslink to 8-oxoguanine DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*C*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOI
 
 | | Structure of G42A human 8-oxoguanine glycosylase crosslinked to undamaged G-containing DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*C*CP*AP*GP*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOB
 
 | | Structure of catalytically inactive H270A human 8-oxoguanine glycosylase crosslinked to 8-oxoguanine DNA | | Descriptor: | 5'-D(*G*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*T*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOE
 
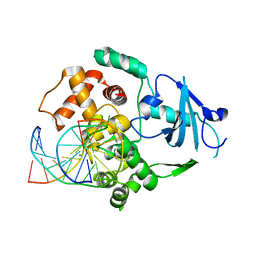 | | Structure of catalytically inactive G42A human 8-oxoguanine glycosylase complexed to 8-oxoguanine DNA | | Descriptor: | 5'-D(*G*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*G*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOH
 
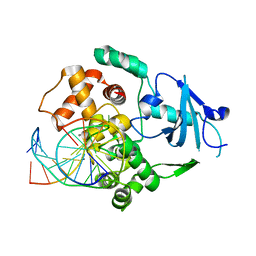 | | Structure of catalytically inactive Q315A human 8-oxoguanine glycosylase complexed to 8-oxoguanine DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOZ
 
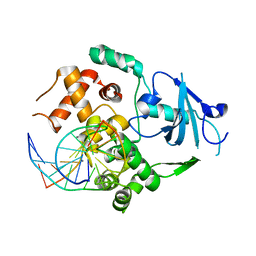 | | Structure of Q315F human 8-oxoguanine glycosylase distal crosslink to 8-oxoguanine DNA | | Descriptor: | 5'-D(*G*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*G*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
