5Q1N
 
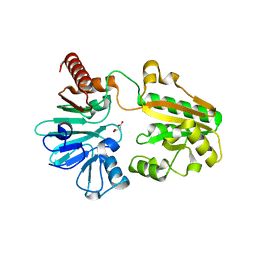 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000295a | | Descriptor: | DNA cross-link repair 1A protein, MALONATE ION, NICKEL (II) ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q22
 
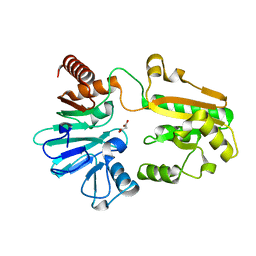 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000728a | | Descriptor: | DNA cross-link repair 1A protein, MALONATE ION, NICKEL (II) ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q2I
 
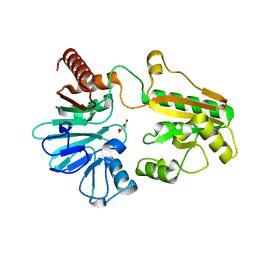 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 10) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q2Y
 
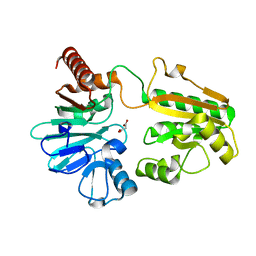 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 26) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q3E
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 42) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q3T
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 57) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5Q4A
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A after initial refinement with no ligand modelled (structure 75) | | Descriptor: | DCLRE1A, MALONATE ION, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5R69
 
 | | XChem group deposition -- Crystal Structure of human YEATS4 in complex with XS038644e | | Descriptor: | 1,2-ETHANEDIOL, YEATS domain-containing protein 4, ~{N}-(5-oxidanylidene-7,8-dihydro-6~{H}-naphthalen-2-yl)ethanamide | | Authors: | Raux, B, Krojer, T, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Huber, K.V.M. | | Deposit date: | 2020-02-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | XChem group deposition
To Be Published
|
|
4KSD
 
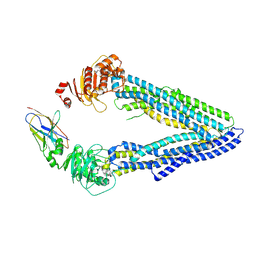 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A, R2 protein | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (4.1001 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KSC
 
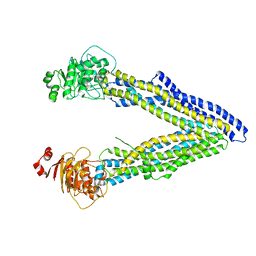 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KSB
 
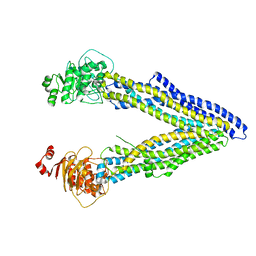 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8001 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7SB3
 
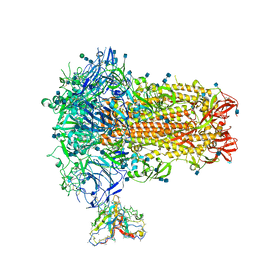 | | Structure of OC43 spike in complex with polyclonal Fab1 (Donor 269) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Ward, A, Bangaru, S, Antanasijevic, A. | | Deposit date: | 2021-09-23 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
5A25
 
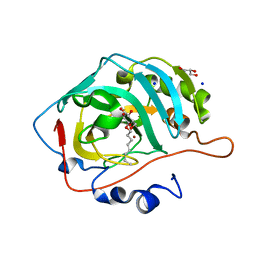 | | Rational engineering of a mesophilic carbonic anhydrase to an extreme halotolerant biocatalyst | | Descriptor: | CARBONIC ANHYDRASE 2, GLYCEROL, SODIUM ION, ... | | Authors: | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | Deposit date: | 2015-05-12 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
4CNW
 
 | | Surface residue engineering of bovine carbonic anhydrase to an extreme halophilic enzyme for potential application in postcombustion CO2 capture | | Descriptor: | CALCIUM ION, CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | Deposit date: | 2014-01-25 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
4CNX
 
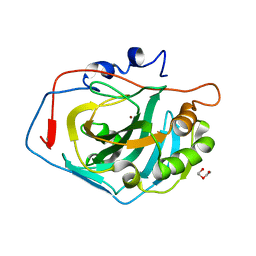 | | Surface residue engineering of bovine carbonic anhydrase to an extreme halophilic enzyme for potential application in postcombustion CO2 capture | | Descriptor: | CARBONIC ANHYDRASE 2, DI(HYDROXYETHYL)ETHER, ZINC ION | | Authors: | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | Deposit date: | 2014-01-25 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
4CNV
 
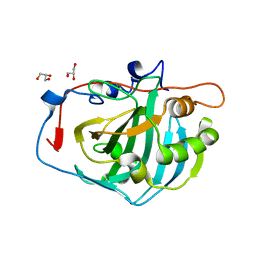 | | Surface residue engineering of bovine carbonic anhydrase to an extreme halophilic enzyme for potential application in postcombustion CO2 capture | | Descriptor: | CARBONIC ANHYDRASE 2, GLYCEROL, ZINC ION | | Authors: | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | Deposit date: | 2014-01-25 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
4CNR
 
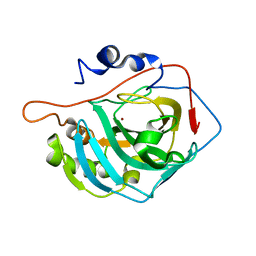 | | Surface residue engineering of bovine carbonic anhydrase to an extreme halophilic enzyme for potential application in postcombustion CO2 capture | | Descriptor: | CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | Deposit date: | 2014-01-24 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
7ZLK
 
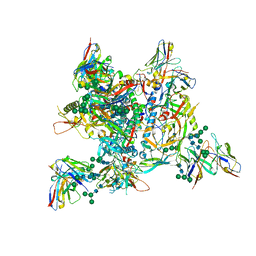 | | AMC009 SOSIPv5.2 in complex with Fabs ACS101 and ACS124 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACS114 heavy chain, ... | | Authors: | van Schooten, J, Ozorowski, G, Ward, A. | | Deposit date: | 2022-04-15 | | Release date: | 2022-09-21 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
Plos Pathog., 18, 2022
|
|
7SBX
 
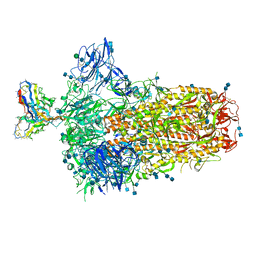 | | Structure of OC43 spike in complex with polyclonal Fab6 (Donor 1051) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-26 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SBY
 
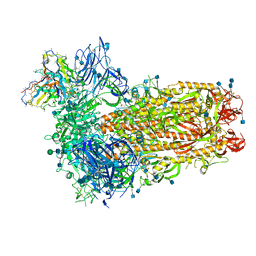 | | Structure of OC43 spike in complex with polyclonal Fab7 (Donor 269) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-26 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SBW
 
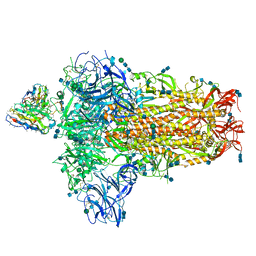 | | Structure of OC43 spike in complex with polyclonal Fab5 (Donor 1051) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-26 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7SBV
 
 | | Structure of OC43 spike in complex with polyclonal Fab4 (Donor 269) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-26 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
7Z3A
 
 | | AMC009 SOSIPv5.2 in complex with Fabs ACS101 and ACS124 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACS101 heavy chain, ... | | Authors: | van Schooten, J, Ward, A. | | Deposit date: | 2022-03-02 | | Release date: | 2022-08-10 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
7SCO
 
 | | Structure of H1 influenza hemagglutinin bound to Fab 310-39G10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 310-39G10 Fab, ... | | Authors: | Torrents de la Pena, A, Ward, A.B. | | Deposit date: | 2021-09-28 | | Release date: | 2022-08-24 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Allelic polymorphism controls autoreactivity and vaccine elicitation of human broadly neutralizing antibodies against influenza virus.
Immunity, 55, 2022
|
|
7SB4
 
 | | Structure of OC43 spike in complex with polyclonal Fab2 (Donor 1412) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-23 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
