2M3L
 
 | |
2M3M
 
 | |
2M8I
 
 | | Structure of Pin1 WW domain | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Luh, L.M, Kirchner, D.K, Loehr, F, Haensel, R, Doetsch, V. | | Deposit date: | 2013-05-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular crowding drives active Pin1 into nonspecific complexes with endogenous proteins prior to substrate recognition.
J.Am.Chem.Soc., 135, 2013
|
|
4PV8
 
 | | Crystal Structure of H2Kb-Q600F complex | | Descriptor: | ACETATE ION, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Twist, K.-A, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-15 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural and functional correlates of enhanced antiviral immunity generated by heteroclitic CD8 T cell epitopes.
J.Immunol., 192, 2014
|
|
4PV9
 
 | | Crystal Structure of H2Kb-Q600V complex | | Descriptor: | ACETATE ION, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Twist, K.-A, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-15 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional correlates of enhanced antiviral immunity generated by heteroclitic CD8 T cell epitopes.
J.Immunol., 192, 2014
|
|
4QRS
 
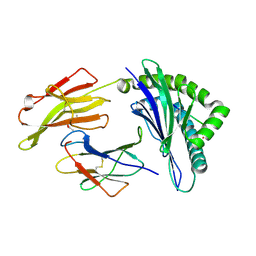 | | Crystal Structure of HLA B*0801 in complex with ELK_IYM, ELKRKMIYM | | Descriptor: | ACETATE ION, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Twist, K.-A, Rossjohn, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-12-10 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular imprint of exposure to naturally occurring genetic variants of human cytomegalovirus on the T cell repertoire.
Sci Rep, 4, 2014
|
|
4PG2
 
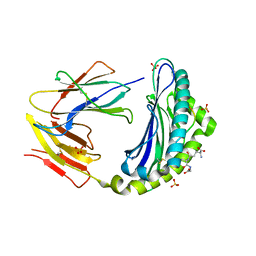 | |
3P5O
 
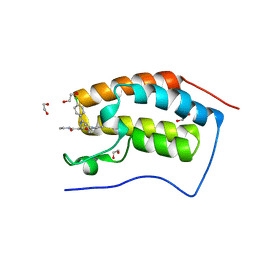 | | Crystal Structure of the First Bromodomain of Human Brd4 in complex with IBET inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(4S)-6-(4-chlorophenyl)-8-methoxy-1-methyl-4H-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-N-ethylacetamide, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2010-10-09 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Suppression of inflammation by a synthetic histone mimic
Nature, 468, 2010
|
|
1REG
 
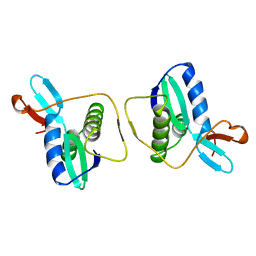 | |
1EBP
 
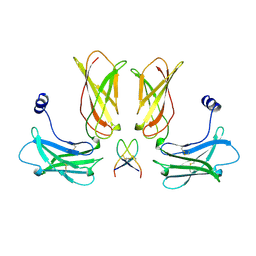 | |
1Q5X
 
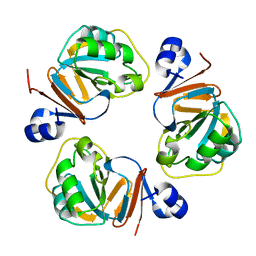 | | Structure of OF RRAA (MENG), a protein inhibitor of RNA processing | | Descriptor: | REGULATOR OF RNASE E ACTIVITY A | | Authors: | Monzingo, A.F, Gao, J, Qiu, J, Georgiou, G, Robertus, J.D. | | Deposit date: | 2003-08-11 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray Structure of Escherichia coli RraA (MenG), A Protein Inhibitor of RNA Processing.
J.Mol.Biol., 332, 2003
|
|
2OKW
 
 | |
2OKY
 
 | |
3T98
 
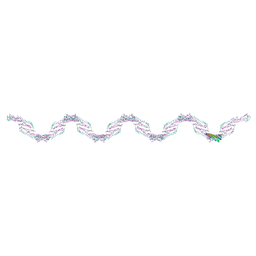 | |
4K2M
 
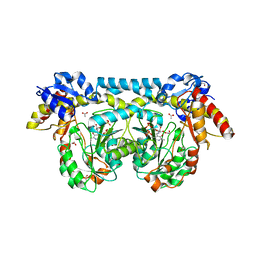 | | Crystal structure of ntda from bacillus subtilis in complex with the plp external aldimine adduct with kanosamine-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 3-deoxy-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-O-phosphono-alpha-D-gluco pyranose, ACETATE ION, ... | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2013-04-09 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Structure of NtdA, a Sugar Aminotransferase Involved in the Kanosamine Biosynthetic Pathway in Bacillus subtilis, Reveals a New Subclass of Aminotransferases.
J.Biol.Chem., 288, 2013
|
|
4K2B
 
 | |
4K2I
 
 | | Crystal structure of ntda from bacillus subtilis with bound cofactor pmp | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ACETATE ION, ... | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2013-04-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.225 Å) | | Cite: | The Structure of NtdA, a Sugar Aminotransferase Involved in the Kanosamine Biosynthetic Pathway in Bacillus subtilis, Reveals a New Subclass of Aminotransferases.
J.Biol.Chem., 288, 2013
|
|
4KBC
 
 | |
4KZ6
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 13 ((2R,6R)-6-methyl-1-(3-sulfanylpropanoyl)piperidine-2-carboxylic acid) | | Descriptor: | (2R,6R)-6-methyl-1-(3-sulfanylpropanoyl)piperidine-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZA
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 48 (3-(cyclopropylsulfamoyl)thiophene-2-carboxylic acid) | | Descriptor: | 3-(cyclopropylsulfamoyl)thiophene-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ7
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 16 ((1R,4S)-4,7,7-trimethyl-3-oxo-2-oxabicyclo[2.2.1]heptane-1-carboxylic acid) | | Descriptor: | (1R,4S)-4,7,7-trimethyl-3-oxo-2-oxabicyclo[2.2.1]heptane-1-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ9
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 41 ((4R,4aS,8aS)-4-phenyldecahydroquinolin-4-ol) | | Descriptor: | (4R,4aS,8aS)-4-phenyldecahydroquinolin-4-ol, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ3
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 44 (5-chloro-3-sulfamoylthiophene-2-carboxylic acid) | | Descriptor: | 5-chloro-3-sulfamoylthiophene-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZB
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 50 (N-(methylsulfonyl)-N-phenyl-alanine) | | Descriptor: | Beta-lactamase, N-(methylsulfonyl)-N-phenyl-D-alanine, N-(methylsulfonyl)-N-phenyl-L-alanine | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ8
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 20 (1,3-diethyl-2-thioxodihydropyrimidine-4,6(1H,5H)-dione) | | Descriptor: | 1,3-diethyl-2-thioxodihydropyrimidine-4,6(1H,5H)-dione, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
